4MS6
 
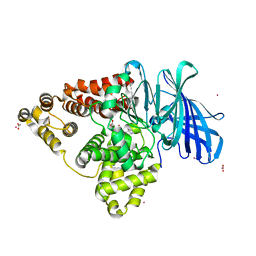 | | Human Leukotriene A4 Hydrolase in complex with Pro-Gly-Pro analogue | | Descriptor: | 1-{4-oxo-4-[(2S)-pyrrolidin-2-yl]butanoyl}-L-proline, ACETIC ACID, Leukotriene A-4 hydrolase, ... | | Authors: | Stsiapanava, A, Rinaldo-Matthis, A, Haeggstrom, J.Z. | | Deposit date: | 2013-09-18 | | Release date: | 2014-03-12 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (1.72 Å) | | Cite: | Binding of Pro-Gly-Pro at the active site of leukotriene A4 hydrolase/aminopeptidase and development of an epoxide hydrolase selective inhibitor.
Proc.Natl.Acad.Sci.USA, 111, 2014
|
|
7KID
 
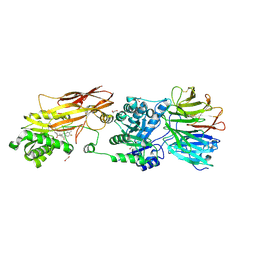 | | PRMT5:MEP50 Complexed with 5,5-Bicyclic Inhibitor Compound 72 | | Descriptor: | (1S,2R,3aR,4S,6aR)-4-[(2-amino-3,5-difluoroquinolin-7-yl)methyl]-2-(4-amino-5-fluoro-7H-pyrrolo[2,3-d]pyrimidin-7-yl)hexahydropentalene-1,6a(1H)-diol, 1,2-ETHANEDIOL, Methylosome protein 50, ... | | Authors: | Palte, R.L. | | Deposit date: | 2020-10-23 | | Release date: | 2021-04-21 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | The Discovery of Two Novel Classes of 5,5-Bicyclic Nucleoside-Derived PRMT5 Inhibitors for the Treatment of Cancer.
J.Med.Chem., 64, 2021
|
|
7KIC
 
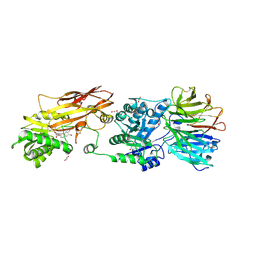 | | PRMT5:MEP50 Complexed with 5,5-Bicyclic Inhibitor Compound 34 | | Descriptor: | (2R,3R,3aS,6S,6aR)-6-[(2-amino-3-bromoquinolin-7-yl)oxy]-2-(4-methyl-7H-pyrrolo[2,3-d]pyrimidin-7-yl)hexahydro-3aH-cyclopenta[b]furan-3,3a-diol, 1,2-ETHANEDIOL, Methylosome protein 50, ... | | Authors: | Palte, R.L. | | Deposit date: | 2020-10-23 | | Release date: | 2021-04-21 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (2.43 Å) | | Cite: | The Discovery of Two Novel Classes of 5,5-Bicyclic Nucleoside-Derived PRMT5 Inhibitors for the Treatment of Cancer.
J.Med.Chem., 64, 2021
|
|
7KIB
 
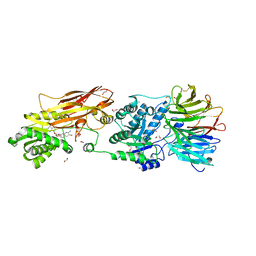 | | PRMT5:MEP50 Complexed with 5,5-Bicyclic Inhibitor Compound 4 | | Descriptor: | (2R,3R,3aS,6S,6aR)-6-[(2-amino-3-bromoquinolin-7-yl)oxy]-2-(4-amino-7H-pyrrolo[2,3-d]pyrimidin-7-yl)hexahydro-3aH-cyclopenta[b]furan-3,3a-diol, 1,2-ETHANEDIOL, CHLORIDE ION, ... | | Authors: | Palte, R.L, Hayes, R.P. | | Deposit date: | 2020-10-23 | | Release date: | 2021-04-21 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (2.52 Å) | | Cite: | The Discovery of Two Novel Classes of 5,5-Bicyclic Nucleoside-Derived PRMT5 Inhibitors for the Treatment of Cancer.
J.Med.Chem., 64, 2021
|
|
4TTH
 
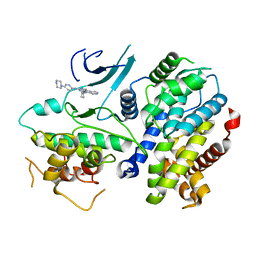 | | Crystal structure of a CDK6/Vcyclin complex with inhibitor bound | | Descriptor: | 9-cyclopentyl-N-(5-piperazin-1-ylpyridin-2-yl)pyrido[4,5]pyrrolo[1,2-d]pyrimidin-2-amine, Cyclin homolog, Cyclin-dependent kinase 6 | | Authors: | Piper, D.E, Walker, N, Wang, Z. | | Deposit date: | 2014-06-20 | | Release date: | 2014-08-06 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (2.9 Å) | | Cite: | Discovery of AMG 925, a FLT3 and CDK4 dual kinase inhibitor with preferential affinity for the activated state of FLT3.
J.Med.Chem., 57, 2014
|
|
1T0Z
 
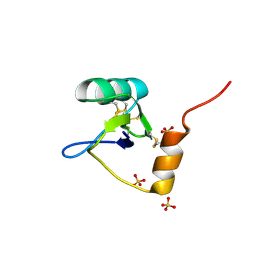 | | Structure of an Excitatory Insect-specific Toxin with an Analgesic Effect on Mammalian from Scorpion Buthus martensii Karsch | | Descriptor: | SULFATE ION, insect neurotoxin | | Authors: | Li, C, Guan, R.-J, Xiang, Y, Zhang, Y, Wang, D.-C. | | Deposit date: | 2004-04-14 | | Release date: | 2004-12-28 | | Last modified: | 2011-07-13 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Structure of an excitatory insect-specific toxin with an analgesic effect on mammals from the scorpion Buthus martensii Karsch.
Acta Crystallogr.,Sect.D, 61, 2005
|
|
4IEH
 
 | |
6N05
 
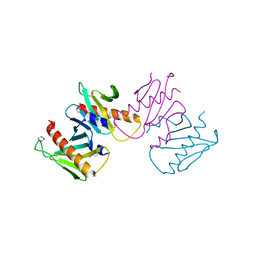 | | Structure of anti-crispr protein, AcrIIC2 | | Descriptor: | AcrIIC2 | | Authors: | Shah, M, Thavalingham, A, Maxwell, K.L, Moraes, T.F. | | Deposit date: | 2018-11-06 | | Release date: | 2019-06-05 | | Last modified: | 2020-01-08 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Inhibition of CRISPR-Cas9 ribonucleoprotein complex assembly by anti-CRISPR AcrIIC2.
Nat Commun, 10, 2019
|
|
6WAA
 
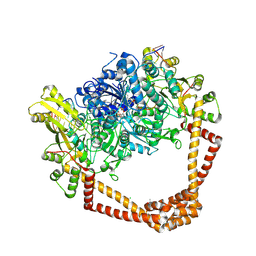 | | K. pneumoniae Topoisomerase IV (ParE-ParC) in complex with DNA and compound 34 (7-[(1S,5R)-1-amino-3-azabicyclo[3.1.0]hexan-3-yl]-4-(aminomethyl)-1-cyclopropyl-3,6-difluoro-8-methylquinolin-2(1H)-one) | | Descriptor: | (4S)-2-METHYL-2,4-PENTANEDIOL, 7-[(1S,5R)-1-amino-3-azabicyclo[3.1.0]hexan-3-yl]-4-(aminomethyl)-1-cyclopropyl-3,6-difluoro-8-methylquinolin-2(1H)-one, CHLORIDE ION, ... | | Authors: | Noeske, J, Shu, W, Bellamacina, C. | | Deposit date: | 2020-03-24 | | Release date: | 2020-07-15 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (3.2 Å) | | Cite: | Topoisomerase Inhibitors Addressing Fluoroquinolone Resistance in Gram-Negative Bacteria.
J.Med.Chem., 63, 2020
|
|
3E9T
 
 | | Crystal structure of Apo-form Calx CBD1 domain | | Descriptor: | CALCIUM ION, Na/Ca exchange protein | | Authors: | Wu, M, Zheng, L. | | Deposit date: | 2008-08-23 | | Release date: | 2009-09-01 | | Last modified: | 2024-05-22 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Crystal structures of progressive Ca2+ binding states of the Ca2+ sensor Ca2+ binding domain 1 (CBD1) from the CALX Na+/Ca2+ exchanger reveal incremental conformational transitions.
J.Biol.Chem., 285, 2010
|
|
7ZKS
 
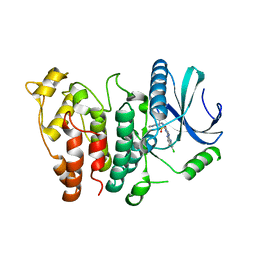 | | SRPK1 IN COMPLEX WITH INHIBITOR | | Descriptor: | CHLORIDE ION, N-[3-[[[2-(6-chloranyl-5-fluoranyl-1H-benzimidazol-2-yl)pyrimidin-4-yl]amino]methyl]pyridin-2-yl]-N-methyl-methanesulfonamide, SRSF protein kinase 1 | | Authors: | Graedler, U. | | Deposit date: | 2022-04-13 | | Release date: | 2023-02-22 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (2.28 Å) | | Cite: | MSC-1186, a Highly Selective Pan-SRPK Inhibitor Based on an Exceptionally Decorated Benzimidazole-Pyrimidine Core.
J.Med.Chem., 66, 2023
|
|
7ZKX
 
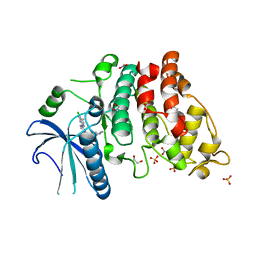 | | SRPK2 IN COMPLEX WITH INHIBITOR | | Descriptor: | 1,2-ETHANEDIOL, ACETATE ION, N-[3-[[[2-(6-chloranyl-5-fluoranyl-1H-benzimidazol-2-yl)pyrimidin-4-yl]amino]methyl]pyridin-2-yl]-N-methyl-methanesulfonamide, ... | | Authors: | Graedler, U. | | Deposit date: | 2022-04-13 | | Release date: | 2023-02-22 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (2.06 Å) | | Cite: | MSC-1186, a Highly Selective Pan-SRPK Inhibitor Based on an Exceptionally Decorated Benzimidazole-Pyrimidine Core.
J.Med.Chem., 66, 2023
|
|
4IJV
 
 | | Crystal structure of 11b-HSD1 double mutant (L262R, F278E) in complex with 3-[1-(4-chlorophenyl)cyclopropyl]-8-(2-fluorophenoxy)[1,2,4]triazolo[4,3-a]pyridine | | Descriptor: | 3-[1-(4-chlorophenyl)cyclopropyl]-8-(2-fluorophenoxy)[1,2,4]triazolo[4,3-a]pyridine, Corticosteroid 11-beta-dehydrogenase isozyme 1, NADP NICOTINAMIDE-ADENINE-DINUCLEOTIDE PHOSPHATE | | Authors: | Sheriff, S. | | Deposit date: | 2012-12-23 | | Release date: | 2014-06-11 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (2.35 Å) | | Cite: | Optimization of 1,2,4-Triazolopyridines as Inhibitors of Human 11 beta-Hydroxysteroid Dehydrogenase Type 1 (11 beta-HSD-1).
ACS Med Chem Lett, 5, 2014
|
|
9ERY
 
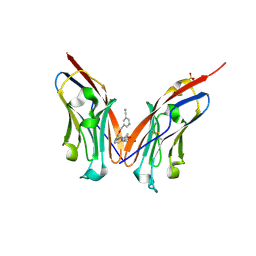 | | Co-crystal strucutre of PD-L1 with low molecular weight inhibitor | | Descriptor: | 5-[[5-[[2-[bis(fluoranyl)methyl]-3-(2,3-dihydro-1,4-benzodioxin-6-yl)phenyl]methoxy]-2-[(2-hydroxyethylamino)methyl]phenoxy]methyl]pyridine-3-carbonitrile, Programmed cell death 1 ligand 1, SULFATE ION | | Authors: | Plewka, J, Magiera-Mularz, K, Zhang, W. | | Deposit date: | 2024-03-25 | | Release date: | 2024-07-24 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | Design, synthesis, and evaluation of antitumor activity of 2-arylmethoxy-4-(2-fluoromethyl-biphenyl-3-ylmethoxy) benzylamine derivatives as PD-1/PD-L1 inhibitors
Eur.J.Med.Chem., 2024
|
|
1THF
 
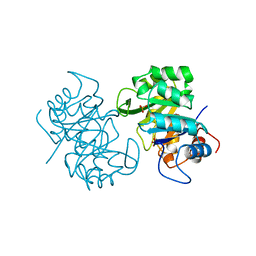 | |
4IJW
 
 | | Crystal structure of 11b-HSD1 double mutant (L262R, F278E) in complex with 3-[1-(4-chlorophenyl)cyclopropyl]-8-cyclopropyl[1,2,4]triazolo[4,3-a]pyridine | | Descriptor: | 3-[1-(4-chlorophenyl)cyclopropyl]-8-cyclopropyl[1,2,4]triazolo[4,3-a]pyridine, Corticosteroid 11-beta-dehydrogenase isozyme 1, NADP NICOTINAMIDE-ADENINE-DINUCLEOTIDE PHOSPHATE | | Authors: | Sheriff, S. | | Deposit date: | 2012-12-23 | | Release date: | 2014-06-11 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (2.35 Å) | | Cite: | Optimization of 1,2,4-Triazolopyridines as Inhibitors of Human 11 beta-Hydroxysteroid Dehydrogenase Type 1 (11 beta-HSD-1).
ACS Med Chem Lett, 5, 2014
|
|
4IJU
 
 | | Crystal structure of 11b-HSD1 double mutant (L262R, F278E) in complex with (1S,4S)-4-[8-(2-fluorophenoxy)[1,2,4]triazolo[4,3-a]pyridin-3-yl]bicyclo[2.2.1]heptan-1-ol | | Descriptor: | (1s,4s)-4-[8-(2-fluorophenoxy)[1,2,4]triazolo[4,3-a]pyridin-3-yl]bicyclo[2.2.1]heptan-1-ol, CHLORIDE ION, Corticosteroid 11-beta-dehydrogenase isozyme 1, ... | | Authors: | Sheriff, S. | | Deposit date: | 2012-12-23 | | Release date: | 2014-06-11 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (2.35 Å) | | Cite: | Optimization of 1,2,4-Triazolopyridines as Inhibitors of Human 11 beta-Hydroxysteroid Dehydrogenase Type 1 (11 beta-HSD-1).
ACS Med Chem Lett, 5, 2014
|
|
4L02
 
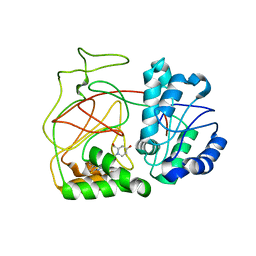 | | Crystal Structure of SphK1 with inhibitor | | Descriptor: | (2R,4S)-1-[2-(4-{[4-(3,4-dichlorophenyl)-1,3-thiazol-2-yl]amino}phenyl)ethyl]-2-(hydroxymethyl)piperidin-4-ol, Sphingosine kinase 1 | | Authors: | Min, X, Walker, N, Wang, Z. | | Deposit date: | 2013-05-30 | | Release date: | 2013-07-24 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (2.75 Å) | | Cite: | Structure guided design of a series of sphingosine kinase (SphK) inhibitors.
Bioorg.Med.Chem.Lett., 23, 2013
|
|
8WB2
 
 | | Heme-bound Arabidopsis thaliana temperature-induced lipocalin | | Descriptor: | PROTOPORPHYRIN IX CONTAINING FE, Temperature-induced lipocalin-1 | | Authors: | Dong, C, Liu, L. | | Deposit date: | 2023-09-08 | | Release date: | 2023-12-27 | | Last modified: | 2024-01-03 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Crystallographic and functional studies of a plant temperature-induced lipocalin.
Biochim Biophys Acta Gen Subj, 1868, 2023
|
|
3GZ9
 
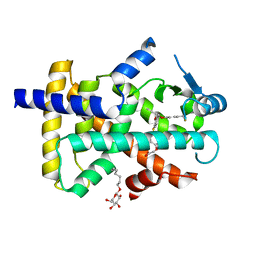 | | Crystal Structure of Peroxisome Proliferator-Activated Receptor Delta (PPARd) in Complex with a Full Agonist | | Descriptor: | (2,3-dimethyl-4-{[2-(prop-2-yn-1-yloxy)-4-{[4-(trifluoromethyl)phenoxy]methyl}phenyl]sulfanyl}phenoxy)acetic acid, Peroxisome proliferator-activated receptor delta, heptyl beta-D-glucopyranoside | | Authors: | Wang, Z, Sudom, A, Walker, N.P. | | Deposit date: | 2009-04-06 | | Release date: | 2009-06-30 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Identification of a PPARdelta agonist with partial agonistic activity on PPARgamma.
Bioorg.Med.Chem.Lett., 19, 2009
|
|
3H0A
 
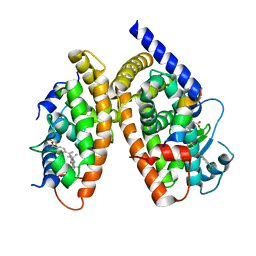 | | Crystal Structure of Peroxisome Proliferator-Activated Receptor Gamma (PPARg) and Retinoic Acid Receptor Alpha (RXRa) in Complex with 9-cis Retinoic Acid, Co-activator Peptide, and a Partial Agonist | | Descriptor: | 4-[1-(3,5,5,8,8-pentamethyl-5,6,7,8-tetrahydronaphthalen-2-yl)ethenyl]benzoic acid, Nuclear receptor coactivator 1, Co-activator Peptide, ... | | Authors: | Wang, Z, Sudom, A, Walker, N.P. | | Deposit date: | 2009-04-08 | | Release date: | 2009-06-09 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Identification of a PPARdelta agonist with partial agonistic activity on PPARgamma.
Bioorg.Med.Chem.Lett., 19, 2009
|
|
6W7F
 
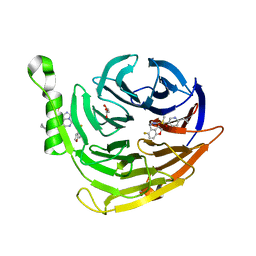 | | Structure of EED bound to inhibitor 5285 | | Descriptor: | 8-(6-cyclopropylpyridin-3-yl)-N-[(5-fluoro-2,3-dihydro-1-benzofuran-4-yl)methyl]-1-(methylsulfonyl)imidazo[1,5-c]pyrimidin-5-amine, GLYCEROL, Polycomb protein EED | | Authors: | Petrunak, E.M, Stuckey, J.A. | | Deposit date: | 2020-03-19 | | Release date: | 2020-07-08 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | EEDi-5285: An Exceptionally Potent, Efficacious, and Orally Active Small-Molecule Inhibitor of Embryonic Ectoderm Development.
J.Med.Chem., 63, 2020
|
|
6W7G
 
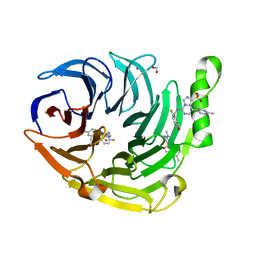 | | Structure of EED bound to inhibitor 1056 | | Descriptor: | 8-(2,6-dimethylpyridin-3-yl)-N-[(5-fluoro-2,3-dihydro-1-benzofuran-4-yl)methyl]-1-(methylsulfonyl)imidazo[1,5-c]pyrimidin-5-amine, FORMIC ACID, Polycomb protein EED, ... | | Authors: | Petrunak, E.M, Stuckey, J.A. | | Deposit date: | 2020-03-19 | | Release date: | 2020-07-08 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (1.85 Å) | | Cite: | EEDi-5285: An Exceptionally Potent, Efficacious, and Orally Active Small-Molecule Inhibitor of Embryonic Ectoderm Development.
J.Med.Chem., 63, 2020
|
|
6IFO
 
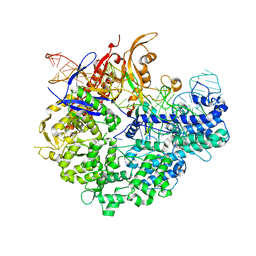 | | Crystal structure of AcrIIA2-SpyCas9-sgRNA ternary complex | | Descriptor: | AcrIIA2, CRISPR-associated endonuclease Cas9/Csn1, RNA (99-MER) | | Authors: | Liu, L, Wang, Y. | | Deposit date: | 2018-09-20 | | Release date: | 2019-01-23 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (3.313 Å) | | Cite: | Phage AcrIIA2 DNA Mimicry: Structural Basis of the CRISPR and Anti-CRISPR Arms Race.
Mol. Cell, 73, 2019
|
|
8X6B
 
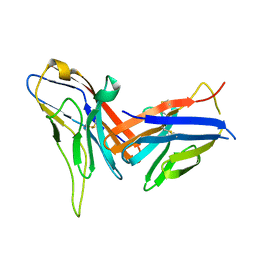 | | Crystal structure of immune receptor PVRIG in complex with ligand Nectin-2 | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, Nectin-2, Transmembrane protein PVRIG | | Authors: | Hu, S.T, Han, P, Wang, H, Qi, J.X. | | Deposit date: | 2023-11-21 | | Release date: | 2024-04-24 | | Last modified: | 2024-07-24 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Structural basis for the immune recognition and selectivity of the immune receptor PVRIG for ligand Nectin-2.
Structure, 32, 2024
|
|
