8TKO
 
 | | KS-AT core of 6-deoxyerythronolide B synthase (DEBS) Module 3 crosslinked with its translocation ACP partner of Module 2 | | Descriptor: | EryAI, EryAII | | Authors: | Cogan, D.P, Soohoo, A.M, Chen, M, Brodsky, K.L, Liu, Y, Khosla, C. | | Deposit date: | 2023-07-25 | | Release date: | 2024-07-31 | | Last modified: | 2025-06-11 | | Method: | ELECTRON MICROSCOPY (3.05 Å) | | Cite: | Structural basis for intermodular communication in assembly-line polyketide biosynthesis.
Nat.Chem.Biol., 21, 2025
|
|
6MZD
 
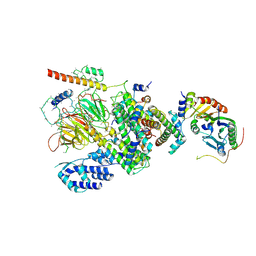 | | Human TFIID Lobe A canonical | | Descriptor: | TATA-box-binding protein, Transcription initiation factor TFIID subunit 1, Transcription initiation factor TFIID subunit 10, ... | | Authors: | Patel, A.B, Louder, R.K, Greber, B.J, Grunberg, S, Luo, J, Fang, J, Liu, Y, Ranish, J, Hahn, S, Nogales, E. | | Deposit date: | 2018-11-05 | | Release date: | 2018-11-28 | | Last modified: | 2025-05-28 | | Method: | ELECTRON MICROSCOPY (9.8 Å) | | Cite: | Structure of human TFIID and mechanism of TBP loading onto promoter DNA.
Science, 362, 2018
|
|
8TGV
 
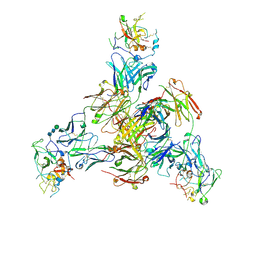 | | CryoEM structure of Fab HC84.26-HCV E2 complex | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, HC84.26 Heavy chain, ... | | Authors: | Shahid, S, Liqun, J, LIu, Y, Hasan, S.S, Mariuzza, R.A. | | Deposit date: | 2023-07-13 | | Release date: | 2024-07-24 | | Last modified: | 2025-07-09 | | Method: | ELECTRON MICROSCOPY (3.75 Å) | | Cite: | Cryo-EM structures of HCV E2 glycoprotein bound to neutralizing and non-neutralizing antibodies determined using bivalent Fabs as fiducial markers.
Commun Biol, 8, 2025
|
|
8TGZ
 
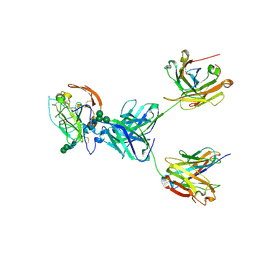 | | CryoEM structure of neutralizing antibody HC84.26 in complex with Hepatitis C virus envelope glycoprotein E2 | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, HC84.26 Heavy chain, ... | | Authors: | Shahid, S, Liqun, J, Liu, Y, Hasan, S.S, Mariuzza, R.A. | | Deposit date: | 2023-07-13 | | Release date: | 2024-07-24 | | Last modified: | 2025-07-09 | | Method: | ELECTRON MICROSCOPY (3.78 Å) | | Cite: | Cryo-EM structures of HCV E2 glycoprotein bound to neutralizing and non-neutralizing antibodies determined using bivalent Fabs as fiducial markers.
Commun Biol, 8, 2025
|
|
8THZ
 
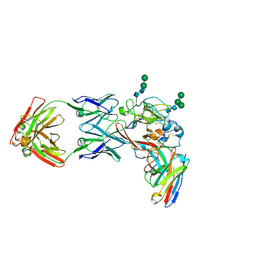 | | CryoEM structure of neutralizing antibodies CBH-7 and HC84.26 in complex with Hepatitis C virus envelope glycoprotein E2 | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-3)-2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, CBH-7 Heavy chain, ... | | Authors: | Shahid, S, Jiang, L, Liu, Y, Hasan, S.S, Mariuzza, R.A. | | Deposit date: | 2023-07-18 | | Release date: | 2024-07-24 | | Last modified: | 2025-07-09 | | Method: | ELECTRON MICROSCOPY (3.25 Å) | | Cite: | Cryo-EM structures of HCV E2 glycoprotein bound to neutralizing and non-neutralizing antibodies determined using bivalent Fabs as fiducial markers.
Commun Biol, 8, 2025
|
|
8TJO
 
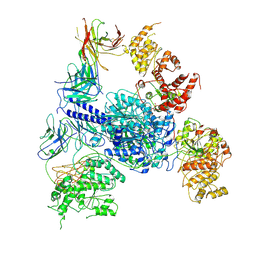 | | Crosslinked 6-deoxyerythronolide B synthase (DEBS) Module 1 in complex with antibody fragment 1B2: Crosslinked Intra-State 1 | | Descriptor: | Antibody Fragment 1B2, Heavy Chain, Light Chain, ... | | Authors: | Cogan, D.P, Soohoo, A.M, Chen, M, Brodsky, K.L, Liu, Y, Khosla, C. | | Deposit date: | 2023-07-23 | | Release date: | 2024-07-24 | | Last modified: | 2025-06-11 | | Method: | ELECTRON MICROSCOPY (3.61 Å) | | Cite: | Structural basis for intermodular communication in assembly-line polyketide biosynthesis.
Nat.Chem.Biol., 21, 2025
|
|
8TPX
 
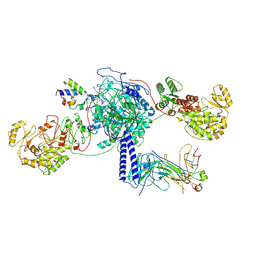 | | Crosslinked 6-deoxyerythronolide B synthase (DEBS) Module 3 in complex with antibody fragment 1B2: trans-oriented 1B2 and ACP | | Descriptor: | Antibody Fragment 1B2, Heavy Chain, Light Chain, ... | | Authors: | Cogan, D.P, Soohoo, A.M, Chen, M, Brodsky, K.L, Liu, Y, Khosla, C. | | Deposit date: | 2023-08-05 | | Release date: | 2024-08-07 | | Last modified: | 2025-06-11 | | Method: | ELECTRON MICROSCOPY (3.4 Å) | | Cite: | Structural basis for intermodular communication in assembly-line polyketide biosynthesis.
Nat.Chem.Biol., 21, 2025
|
|
8TJP
 
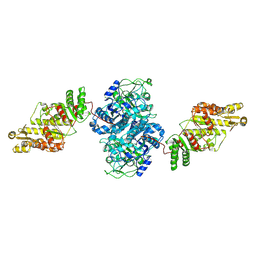 | | KS-AT core of 6-deoxyerythronolide B synthase (DEBS) Module 3 crosslinked with its elongation ACP partner | | Descriptor: | EryAII | | Authors: | Cogan, D.P, Soohoo, A.M, Chen, M, Brodsky, K.L, Liu, Y, Khosla, C. | | Deposit date: | 2023-07-23 | | Release date: | 2024-07-24 | | Last modified: | 2025-06-11 | | Method: | ELECTRON MICROSCOPY (3.71 Å) | | Cite: | Structural basis for intermodular communication in assembly-line polyketide biosynthesis.
Nat.Chem.Biol., 21, 2025
|
|
8TJN
 
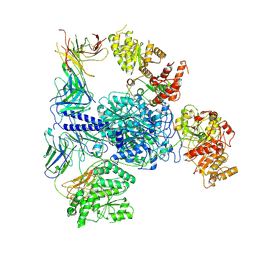 | | Crosslinked 6-deoxyerythronolide B synthase (DEBS) Module 1 in complex with antibody fragment 1B2: Crosslinked State 1 | | Descriptor: | Antibody Fragment 1B2, Heavy Chain, Light Chain, ... | | Authors: | Cogan, D.P, Soohoo, A.M, Chen, M, Brodsky, K.L, Liu, Y, Khosla, C. | | Deposit date: | 2023-07-23 | | Release date: | 2024-07-24 | | Last modified: | 2025-06-11 | | Method: | ELECTRON MICROSCOPY (3.73 Å) | | Cite: | Structural basis for intermodular communication in assembly-line polyketide biosynthesis.
Nat.Chem.Biol., 21, 2025
|
|
8TPW
 
 | | Crosslinked 6-deoxyerythronolide B synthase (DEBS) Module 3 in complex with antibody fragment 1B2: cis-oriented 1B2 and ACP | | Descriptor: | Antibody Fragment 1B2, Heavy Chain, Light Chain, ... | | Authors: | Cogan, D.P, Soohoo, A.M, Chen, M, Brodsky, K.L, Liu, Y, Khosla, C. | | Deposit date: | 2023-08-05 | | Release date: | 2024-08-07 | | Last modified: | 2025-06-11 | | Method: | ELECTRON MICROSCOPY (3.46 Å) | | Cite: | Structural basis for intermodular communication in assembly-line polyketide biosynthesis.
Nat.Chem.Biol., 21, 2025
|
|
6MZL
 
 | | Human TFIID canonical state | | Descriptor: | TATA-box-binding protein, Transcription initiation factor TFIID subunit 1, Transcription initiation factor TFIID subunit 10, ... | | Authors: | Patel, A.B, Louder, R.K, Greber, B.J, Grunberg, S, Luo, J, Fang, J, Liu, Y, Ranish, J, Hahn, S, Nogales, E. | | Deposit date: | 2018-11-05 | | Release date: | 2018-11-28 | | Last modified: | 2024-11-13 | | Method: | ELECTRON MICROSCOPY (23 Å) | | Cite: | Structure of human TFIID and mechanism of TBP loading onto promoter DNA.
Science, 362, 2018
|
|
5A9G
 
 | | Manganese Superoxide Dismutase from Sphingobacterium sp. T2 | | Descriptor: | MANGANESE (II) ION, MANGANESE SUPEROXIDE DISMUTASE | | Authors: | Rashid, G.M.M, Taylor, C.R, Liu, Y, Zhang, X, Rea, D, Fulop, V, Bugg, T.D.H. | | Deposit date: | 2015-07-21 | | Release date: | 2015-08-05 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (1.35 Å) | | Cite: | Identification of Manganese Superoxide Dismutase from Sphingobacterium Sp. T2 as a Novel Bacterial Enzyme for Lignin Oxidation.
Acs Chem.Biol., 10, 2015
|
|
6OAI
 
 | | Crystal structure of P[6] rotavirus vp8* complexed with LNFPI | | Descriptor: | Protease-sensitive outer capsid protein, alpha-L-fucopyranose-(1-2)-beta-D-galactopyranose-(1-3)-2-acetamido-2-deoxy-beta-D-glucopyranose-(1-3)-beta-D-galactopyranose-(1-4)-beta-D-glucopyranose | | Authors: | Xu, S, Liu, Y, Jiang, X, Kennedy, M.A. | | Deposit date: | 2019-03-16 | | Release date: | 2020-03-18 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Molecular basis of P[II] major human rotavirus VP8* domain recognition of histo-blood group antigens.
Plos Pathog., 16, 2020
|
|
6OJ0
 
 | | Cryo-EM reconstruction of Sulfolobus polyhedral virus 1 (SPV1) | | Descriptor: | Structural protein VP4, Uncharacterized protein | | Authors: | Wang, F, Liu, Y, Conway, J.F, Krupovic, M, Prangishvili, D, Egelman, E.H. | | Deposit date: | 2019-04-10 | | Release date: | 2019-10-02 | | Last modified: | 2025-05-14 | | Method: | ELECTRON MICROSCOPY (3.7 Å) | | Cite: | A packing for A-form DNA in an icosahedral virus.
Proc.Natl.Acad.Sci.USA, 116, 2019
|
|
3CXR
 
 | | Crystal structure of gluconate 5-dehydrogase from streptococcus suis type 2 | | Descriptor: | Dehydrogenase with different specificities | | Authors: | Peng, H, Gao, F, Zhang, Q, Liu, Y, Gao, G.F. | | Deposit date: | 2008-04-25 | | Release date: | 2009-03-17 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Structural insight into the catalytic mechanism of gluconate 5-dehydrogenase from Streptococcus suis: Crystal structures of the substrate-free and quaternary complex enzymes.
Protein Sci., 18, 2009
|
|
3CM8
 
 | | A RNA polymerase subunit structure from virus | | Descriptor: | Polymerase acidic protein, peptide from RNA-directed RNA polymerase catalytic subunit | | Authors: | He, X, Zhou, J, Zeng, Z, Ma, J, Zhang, R, Rao, Z, Liu, Y. | | Deposit date: | 2008-03-21 | | Release date: | 2008-07-15 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (2.899 Å) | | Cite: | Crystal structure of the polymerase PAC-PB1N complex from an avian influenza H5N1 virus
Nature, 454, 2008
|
|
8GI2
 
 | | Cryo-EM structure of Natrinema sp. J7-2 Type IV pilus, PilA2 | | Descriptor: | Natrinema pilin, PilA2 | | Authors: | Sonani, R.R, Kreutzberger, M.A.B, Liu, Y, Krupovic, M, Egelman, E.H. | | Deposit date: | 2023-03-13 | | Release date: | 2023-08-16 | | Last modified: | 2025-06-18 | | Method: | ELECTRON MICROSCOPY (3 Å) | | Cite: | The evolution of archaeal flagellar filaments.
Proc.Natl.Acad.Sci.USA, 120, 2023
|
|
9DR1
 
 | | E. coli RNA polymerase consensus volume with a bound fluoride riboswitch in the ligand-bound state | | Descriptor: | DNA (30-MER), DNA (39-MER), DNA-directed RNA polymerase subunit alpha, ... | | Authors: | Porta, J.C, Ellinger, E, Liu, Y, Walter, N.G. | | Deposit date: | 2024-09-24 | | Release date: | 2024-10-09 | | Last modified: | 2025-02-12 | | Method: | ELECTRON MICROSCOPY (3.7 Å) | | Cite: | Structural basis of RNA-mediated regulation of transcriptional pausing.
To Be Published
|
|
9DQH
 
 | | CryoEM structure of Gq-coupled MRGPRD with a new agonist EP-2825 | | Descriptor: | 2-({1-[2-(4-chlorophenyl)-2-methylpropanoyl]piperidin-4-yl}amino)-5,6,7,8-tetrahydroquinazolin-4(3H)-one, Gs-mini-Gq chimera, Guanine nucleotide-binding protein G(I)/G(S)/G(O) subunit gamma-2, ... | | Authors: | Cao, C, Wang, C, Liu, Y, Fay, J.F, Roth, B.L. | | Deposit date: | 2024-09-24 | | Release date: | 2024-12-11 | | Last modified: | 2025-05-28 | | Method: | ELECTRON MICROSCOPY (2.92 Å) | | Cite: | High-affinity agonists reveal recognition motifs for the MRGPRD GPCR.
Cell Rep, 43, 2024
|
|
7TZC
 
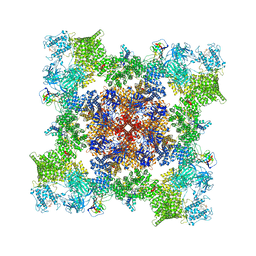 | | A drug and ATP binding site in type 1 ryanodine receptor | | Descriptor: | (2S)-3-(octadecanoyloxy)-2-[(9Z)-octadec-9-enoyloxy]propyl 2-(trimethylammonio)ethyl phosphate, 4-[(7-methoxy-2,3-dihydro-1,4-benzothiazepin-4(5H)-yl)methyl]benzoic acid, ADENOSINE-5'-TRIPHOSPHATE, ... | | Authors: | Melville, Z, Dridi, H, Yuan, Q, Reiken, S, Anetta, W, Liu, Y, Clarke, O.B, Marks, A.R. | | Deposit date: | 2022-02-15 | | Release date: | 2022-05-18 | | Last modified: | 2024-11-13 | | Method: | ELECTRON MICROSCOPY (2.45 Å) | | Cite: | A drug and ATP binding site in type 1 ryanodine receptor.
Structure, 30, 2022
|
|
9DQJ
 
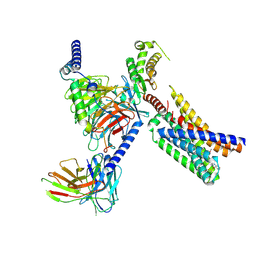 | | CryoEM structure of Gq-coupled MRGPRD with a new agonist EP-3945 | | Descriptor: | 2-({1-[1-(4-methoxyphenyl)cyclopropane-1-carbonyl]piperidin-4-yl}amino)quinazolin-4(3H)-one, Guanine nucleotide-binding protein G(I)/G(S)/G(O) subunit gamma-2, Guanine nucleotide-binding protein G(I)/G(S)/G(T) subunit beta-1, ... | | Authors: | Cao, C, Wang, C, Liu, Y, Fay, J.F, Roth, B.L. | | Deposit date: | 2024-09-24 | | Release date: | 2024-12-11 | | Last modified: | 2025-05-28 | | Method: | ELECTRON MICROSCOPY (2.9 Å) | | Cite: | High-affinity agonists reveal recognition motifs for the MRGPRD GPCR.
Cell Rep, 43, 2024
|
|
7X3O
 
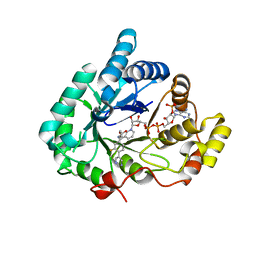 | | Crystal structure of Aldo-keto reductase 1C3 complexed with compound S07054 | | Descriptor: | (2~{R})-2-(3-fluoranyl-4-pyrimidin-5-yl-phenyl)butanoic acid, Aldo-keto reductase family 1 member C3, NADP NICOTINAMIDE-ADENINE-DINUCLEOTIDE PHOSPHATE | | Authors: | Jiang, J, Liu, Y, He, S, Chen, Y, Chu, X, Liu, Y, Guo, Q, Zhao, L, Feng, F, Liu, W, Zhang, X, Fang, P, Sun, H. | | Deposit date: | 2022-03-01 | | Release date: | 2023-03-08 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2.001 Å) | | Cite: | Crystal structure of Aldo-keto reductase 1C3 complexed with compound S07054
To Be Published
|
|
7X3L
 
 | | Crystal structure of Aldo-keto reductase 1C3 complexed with compound S07044 | | Descriptor: | (2~{R})-2-[4-(3-fluoranyl-4-methyl-phenyl)-3-(trifluoromethyl)phenyl]butanoic acid, Aldo-keto reductase family 1 member C3, NADP NICOTINAMIDE-ADENINE-DINUCLEOTIDE PHOSPHATE | | Authors: | Jiang, J, Liu, Y, He, S, Chen, Y, Chu, X, Liu, Y, Guo, Q, Zhao, L, Feng, F, Liu, W, Zhang, X, Fang, P, Sun, H. | | Deposit date: | 2022-03-01 | | Release date: | 2023-03-08 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (1.86 Å) | | Cite: | Development of Biaryl-Containing Aldo-Keto Reductase 1C3 (AKR1C3) Inhibitors for Reversing AKR1C3-Mediated Drug Resistance in Cancer Treatment.
J.Med.Chem., 66, 2023
|
|
7X3M
 
 | | Crystal structure of Aldo-keto reductase 1C3 complexed with compound S07045 | | Descriptor: | (2~{R})-2-[4-[3,5-bis(chloranyl)phenyl]-3-(trifluoromethyl)phenyl]butanoic acid, Aldo-keto reductase family 1 member C3, NADP NICOTINAMIDE-ADENINE-DINUCLEOTIDE PHOSPHATE | | Authors: | Jiang, J, Liu, Y, He, S, Chen, Y, Chu, X, Liu, Y, Guo, Q, Zhao, L, Feng, F, Liu, W, Zhang, X, Fang, P, Sun, H. | | Deposit date: | 2022-03-01 | | Release date: | 2023-03-08 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2.694 Å) | | Cite: | Development of Biaryl-Containing Aldo-Keto Reductase 1C3 (AKR1C3) Inhibitors for Reversing AKR1C3-Mediated Drug Resistance in Cancer Treatment.
J.Med.Chem., 66, 2023
|
|
2H40
 
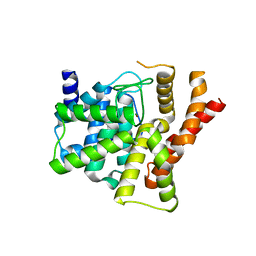 | | Crystal structure of the catalytic domain of unliganded PDE5 | | Descriptor: | MAGNESIUM ION, ZINC ION, cGMP-specific 3',5'-cyclic phosphodiesterase | | Authors: | Wang, H, Liu, Y, Huai, Q, Cai, J, Zoraghi, R, Francis, S.H, Corbin, J.D, Robinson, H, Xin, Z, Lin, G, Ke, H. | | Deposit date: | 2006-05-23 | | Release date: | 2006-06-06 | | Last modified: | 2024-11-13 | | Method: | X-RAY DIFFRACTION (1.85 Å) | | Cite: | Multiple Conformations of Phosphodiesterase-5: Implications for enzyme function and drug development
J.Biol.Chem., 281, 2006
|
|
