7OV3
 
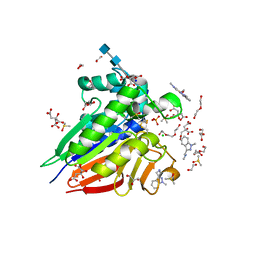 | | Crystal structure of pig purple acid phosphatase in complex with Maybridge fragment CC063346, dimethyl sulfoxide and citrate | | Descriptor: | 1,2-ETHANEDIOL, 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, ... | | Authors: | Feder, D, McGeary, R.P, Guddat, L.W, Schenk, G. | | Deposit date: | 2021-06-14 | | Release date: | 2022-01-12 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Rational Design of Potent Inhibitors of a Metallohydrolase Using a Fragment-Based Approach.
Chemmedchem, 16, 2021
|
|
7OV2
 
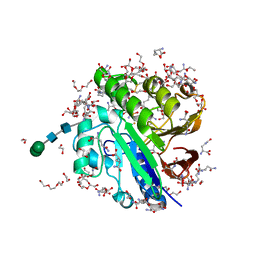 | | Crystal structure of pig purple acid phosphatase in complex with L-glutamine, (poly)ethylene glycol fragments and glycerol | | Descriptor: | 1,2-ETHANEDIOL, CITRIC ACID, FE (III) ION, ... | | Authors: | Feder, D, McGeary, R.P, Guddat, L.W, Schenk, G. | | Deposit date: | 2021-06-14 | | Release date: | 2022-01-12 | | Last modified: | 2024-11-20 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Rational Design of Potent Inhibitors of a Metallohydrolase Using a Fragment-Based Approach.
Chemmedchem, 16, 2021
|
|
7OV8
 
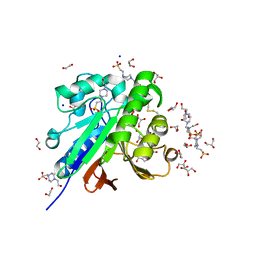 | | Crystal structure of pig purple acid phosphatase in complex with 4-(2-hydroxyethyl)-1-piperazineethanesulfonic acid (HEPES) and glycerol | | Descriptor: | 1,2-ETHANEDIOL, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, 4-(2-HYDROXYETHYL)-1-PIPERAZINE ETHANESULFONIC ACID, ... | | Authors: | Feder, D, McGeary, R.P, Guddat, L.W, Schenk, G. | | Deposit date: | 2021-06-14 | | Release date: | 2022-03-23 | | Last modified: | 2024-10-09 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Rational Design of Potent Inhibitors of a Metallohydrolase Using a Fragment-Based Approach.
Chemmedchem, 16, 2021
|
|
4IZD
 
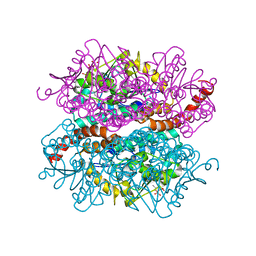 | | Crystal structure of DmdD E121A in complex with MMPA-CoA | | Descriptor: | 3-methylmercaptopropionate-CoA (MMPA-CoA), Enoyl-CoA hydratase/isomerase family protein | | Authors: | Tan, D, Tong, L. | | Deposit date: | 2013-01-29 | | Release date: | 2013-06-05 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Crystal Structure of DmdD, a Crotonase Superfamily Enzyme That Catalyzes the Hydration and Hydrolysis of Methylthioacryloyl-CoA.
Plos One, 8, 2013
|
|
1D70
 
 | |
4KQY
 
 | | Bacillus subtilis yitJ S box/SAM-I riboswitch | | Descriptor: | MAGNESIUM ION, S-ADENOSYLMETHIONINE, YitJ S box/SAM-I riboswitch | | Authors: | Lu, C. | | Deposit date: | 2013-05-15 | | Release date: | 2013-08-07 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (3.02 Å) | | Cite: | SAM recognition and conformational switching mechanism in the Bacillus subtilis yitJ S box/SAM-I riboswitch
J.Mol.Biol., 404, 2010
|
|
4KR6
 
 | |
4IZC
 
 | | Crystal structure of DmdD E121A in complex with MTA-CoA | | Descriptor: | Enoyl-CoA hydratase/isomerase family protein, methylthioacryloyl-CoA | | Authors: | Tan, D, Tong, L. | | Deposit date: | 2013-01-29 | | Release date: | 2013-06-05 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Crystal Structure of DmdD, a Crotonase Superfamily Enzyme That Catalyzes the Hydration and Hydrolysis of Methylthioacryloyl-CoA.
Plos One, 8, 2013
|
|
4KR7
 
 | | Crystal structure of a 4-thiouridine synthetase - RNA complex with bound ATP | | Descriptor: | ADENOSINE-5'-TRIPHOSPHATE, MAGNESIUM ION, Probable tRNA sulfurtransferase, ... | | Authors: | Neumann, P, Ficner, R, Lakomek, K. | | Deposit date: | 2013-05-16 | | Release date: | 2014-04-23 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (3.421 Å) | | Cite: | Crystal structure of a 4-thiouridine synthetase-RNA complex reveals specificity of tRNA U8 modification.
Nucleic Acids Res., 42, 2014
|
|
4IZB
 
 | |
4KR9
 
 | |
7S05
 
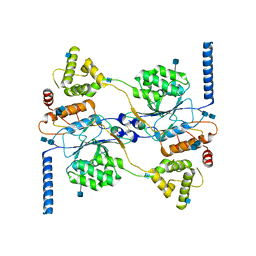 | | Cryo-EM structure of human GlcNAc-1-phosphotransferase A2B2 subcomplex | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, CALCIUM ION, ... | | Authors: | Li, H, Li, H. | | Deposit date: | 2021-08-30 | | Release date: | 2022-03-30 | | Last modified: | 2024-10-16 | | Method: | ELECTRON MICROSCOPY (3.1 Å) | | Cite: | Structure of the human GlcNAc-1-phosphotransferase alpha beta subunits reveals regulatory mechanism for lysosomal enzyme glycan phosphorylation.
Nat.Struct.Mol.Biol., 29, 2022
|
|
7S06
 
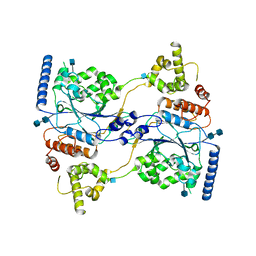 | | Cryo-EM structure of human GlcNAc-1-phosphotransferase A2B2 subcomplex | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, N-acetylglucosamine-1-phosphotransferase subunits alpha/beta | | Authors: | Li, H, Li, H. | | Deposit date: | 2021-08-30 | | Release date: | 2022-03-30 | | Last modified: | 2024-11-06 | | Method: | ELECTRON MICROSCOPY (3.3 Å) | | Cite: | Structure of the human GlcNAc-1-phosphotransferase alpha beta subunits reveals regulatory mechanism for lysosomal enzyme glycan phosphorylation.
Nat.Struct.Mol.Biol., 29, 2022
|
|
7RNG
 
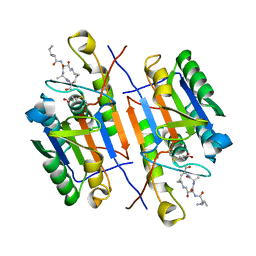 | |
7RNA
 
 | |
7SR2
 
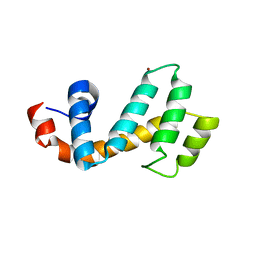 | | Crystal structure of the human SNX25 regulator of G-protein signalling (RGS) domain | | Descriptor: | ACETATE ION, LEUCINE, Sorting nexin-25, ... | | Authors: | Collins, B.M, Paul, B, Weeratunga, S. | | Deposit date: | 2021-11-07 | | Release date: | 2021-11-17 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (2.42 Å) | | Cite: | Structural Predictions of the SNX-RGS Proteins Suggest They Belong to a New Class of Lipid Transfer Proteins.
Front Cell Dev Biol, 10, 2022
|
|
7SR1
 
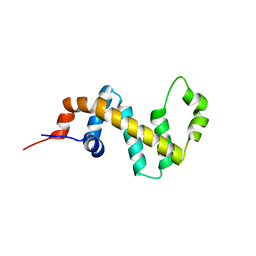 | |
7PV8
 
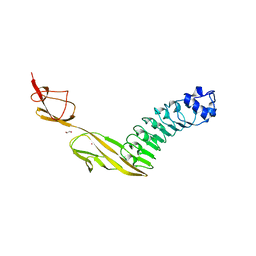 | |
7PV9
 
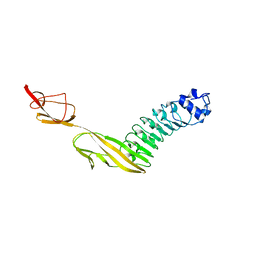 | |
7SSB
 
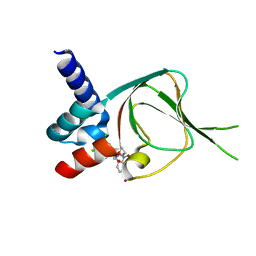 | | Co-structure of PKG1 regulatory domain with compound 33 | | Descriptor: | 4-({(2S,3S)-3-[(1S)-1-(3,5-dichlorophenyl)-2-hydroxyethoxy]-2-phenylpiperidin-1-yl}methyl)-3-nitrobenzoic acid, cGMP-dependent protein kinase 1 | | Authors: | Fischmann, T.O. | | Deposit date: | 2021-11-10 | | Release date: | 2022-08-24 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (1.4 Å) | | Cite: | Optimization and Mechanistic Investigations of Novel Allosteric Activators of PKG1 alpha.
J.Med.Chem., 65, 2022
|
|
8FW0
 
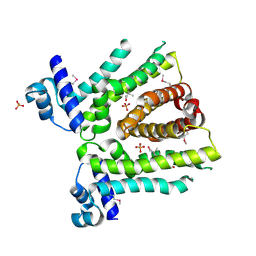 | | MtrR from Neisseria gonorrhoeae bound to beta-Estradiol | | Descriptor: | ESTRADIOL, GLYCEROL, HTH-type transcriptional regulator MtrR, ... | | Authors: | Hooks, G.H, Brennan, R.G. | | Deposit date: | 2023-01-20 | | Release date: | 2024-01-24 | | Last modified: | 2024-11-13 | | Method: | X-RAY DIFFRACTION (2.37 Å) | | Cite: | Hormonal steroids induce multidrug resistance and stress response genes in Neisseria gonorrhoeae by binding to MtrR.
Nat Commun, 15, 2024
|
|
8FW3
 
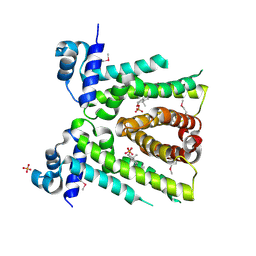 | | MtrR from Neisseria gonorrhoeae bound to Testosterone | | Descriptor: | HTH-type transcriptional regulator MtrR, PHOSPHATE ION, TESTOSTERONE | | Authors: | Hooks, G.H, Brennan, R.G. | | Deposit date: | 2023-01-20 | | Release date: | 2024-01-24 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (2.22 Å) | | Cite: | Hormonal steroids induce multidrug resistance and stress response genes in Neisseria gonorrhoeae by binding to MtrR.
Nat Commun, 15, 2024
|
|
8FW8
 
 | | MtrR from Neisseria gonorrhoeae bound to Progesterone | | Descriptor: | 3-CYCLOHEXYL-1-PROPYLSULFONIC ACID, HTH-type transcriptional regulator MtrR, PHOSPHATE ION, ... | | Authors: | Hooks, G.H, Brennan, R.G. | | Deposit date: | 2023-01-20 | | Release date: | 2024-01-24 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (2.31 Å) | | Cite: | Hormonal steroids induce multidrug resistance and stress response genes in Neisseria gonorrhoeae by binding to MtrR.
Nat Commun, 15, 2024
|
|
8GNJ
 
 | | Human SARM1 bounded with NMN and Nanobody-C6, Conformation 2 | | Descriptor: | BETA-NICOTINAMIDE RIBOSE MONOPHOSPHATE, NAD(+) hydrolase SARM1, Nanobody-C6 | | Authors: | Cai, Y, Zhang, H. | | Deposit date: | 2022-08-24 | | Release date: | 2023-01-18 | | Last modified: | 2024-10-16 | | Method: | ELECTRON MICROSCOPY (3.78 Å) | | Cite: | A conformation-specific nanobody targeting the nicotinamide mononucleotide-activated state of SARM1.
Nat Commun, 13, 2022
|
|
8GNI
 
 | | Human SARM1 bounded with NMN and Nanobody-C6, Conformation 1 | | Descriptor: | BETA-NICOTINAMIDE RIBOSE MONOPHOSPHATE, NAD(+) hydrolase SARM1, Nanobody C6 | | Authors: | Cai, Y, Zhang, H. | | Deposit date: | 2022-08-24 | | Release date: | 2023-01-18 | | Last modified: | 2024-10-23 | | Method: | ELECTRON MICROSCOPY (3.74 Å) | | Cite: | A conformation-specific nanobody targeting the nicotinamide mononucleotide-activated state of SARM1.
Nat Commun, 13, 2022
|
|
