7XI0
 
 | | Crystal structure of CBP bromodomain liganded with CCS150 | | Descriptor: | (6S)-1-(3-chloranyl-4-methoxy-phenyl)-6-[5-(3,5-dimethyl-1,2-oxazol-4-yl)-1-[(3R)-1-methylsulfonylpyrrolidin-3-yl]benzimidazol-2-yl]piperidin-2-one, CREB-binding protein, GLYCEROL | | Authors: | Xu, H, Xiang, Q, Wang, C, Zhang, C, Luo, G, Wu, X, Zhang, Y, Xu, Y. | | Deposit date: | 2022-04-11 | | Release date: | 2022-07-27 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (1.62 Å) | | Cite: | Structural insights revealed by the cocrystal structure of CCS1477 in complex with CBP bromodomain
Biochem.Biophys.Res.Commun., 623, 2022
|
|
7XHE
 
 | | Crystal structure of CBP bromodomain liganded with CCS151 | | Descriptor: | (6S)-6-[5-(3,5-dimethyl-1,2-oxazol-4-yl)-1-[(3R)-1-methylsulfonylpyrrolidin-3-yl]benzimidazol-2-yl]-1-(3-fluoranyl-4-methoxy-phenyl)piperidin-2-one, 1,2-ETHANEDIOL, CREB-binding protein | | Authors: | Xu, H, Xiang, Q, Wang, C, Zhang, C, Luo, G, Wu, X, Zhang, Y, Xu, Y. | | Deposit date: | 2022-04-08 | | Release date: | 2022-07-27 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (1.59 Å) | | Cite: | Structural insights revealed by the cocrystal structure of CCS1477 in complex with CBP bromodomain
Biochem.Biophys.Res.Commun., 623, 2022
|
|
7MDW
 
 | |
7ME7
 
 | |
7MEJ
 
 | |
4GJT
 
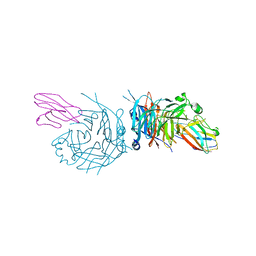 | | complex structure of nectin-4 bound to MV-H | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, Hemagglutinin glycoprotein, Poliovirus receptor-related protein 4 | | Authors: | Zhang, X, Lu, G, Qi, J, Li, Y, He, Y, Xu, X, Shi, J, Zhang, C, Yan, J, Gao, G.F. | | Deposit date: | 2012-08-10 | | Release date: | 2012-10-10 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (3.1001 Å) | | Cite: | Structure of measles virus hemagglutinin bound to its epithelial receptor nectin-4
Nat.Struct.Mol.Biol., 20, 2013
|
|
7XPN
 
 | |
6IJX
 
 | | Crystal Structure of AKR1C1 complexed with meclofenamic acid | | Descriptor: | 2-[(2,6-dichloro-3-methyl-phenyl)amino]benzoic acid, Aldo-keto reductase family 1 member C1, NADP NICOTINAMIDE-ADENINE-DINUCLEOTIDE PHOSPHATE | | Authors: | Zheng, X, Zhao, Y, Zhang, L, Zhang, H, Chen, Y, Hu, X. | | Deposit date: | 2018-10-12 | | Release date: | 2019-10-16 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Screening, synthesis, crystal structure, and molecular basis of 6-amino-4-phenyl-1,4-dihydropyrano[2,3-c]pyrazole-5-carbonitriles as novel AKR1C3 inhibitors.
Bioorg.Med.Chem., 26, 2018
|
|
7N9T
 
 | |
7P1J
 
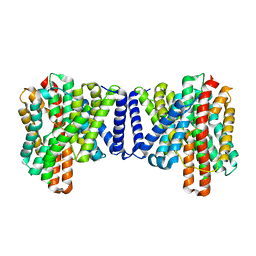 | | Cryo EM structure of bison NHA2 in detergent structure | | Descriptor: | mitochondrial sodium/hydrogen exchanger 9B2 | | Authors: | Matsuoka, R, Fudim, R, Jung, S, Drew, D. | | Deposit date: | 2021-07-01 | | Release date: | 2022-01-26 | | Last modified: | 2022-03-02 | | Method: | ELECTRON MICROSCOPY (3.04 Å) | | Cite: | Structure, mechanism and lipid-mediated remodeling of the mammalian Na + /H + exchanger NHA2.
Nat.Struct.Mol.Biol., 29, 2022
|
|
7P1I
 
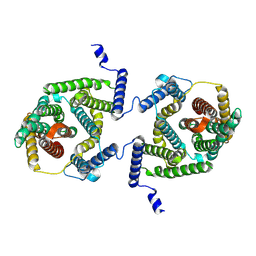 | | Cryo EM structure of bison NHA2 in detergent and N-terminal extension helix | | Descriptor: | mitochondrial sodium/hydrogen exchanger 9B2 | | Authors: | Matsuoka, R, Fudim, R, Jung, S, Drew, D. | | Deposit date: | 2021-07-01 | | Release date: | 2022-01-26 | | Last modified: | 2022-03-02 | | Method: | ELECTRON MICROSCOPY (3.15 Å) | | Cite: | Structure, mechanism and lipid-mediated remodeling of the mammalian Na + /H + exchanger NHA2.
Nat.Struct.Mol.Biol., 29, 2022
|
|
7P1K
 
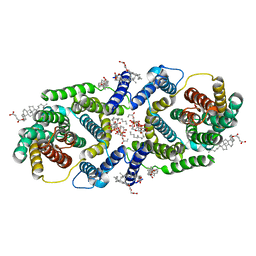 | | Cryo EM structure of bison NHA2 in nano disc structure | | Descriptor: | CHOLESTEROL HEMISUCCINATE, Phosphatidylinositol, mitochondrial sodium/hydrogen exchanger 9B2 | | Authors: | Matsuoka, R, Fudim, R, Jung, S, Drew, D. | | Deposit date: | 2021-07-01 | | Release date: | 2022-01-26 | | Last modified: | 2022-03-02 | | Method: | ELECTRON MICROSCOPY (3.64 Å) | | Cite: | Structure, mechanism and lipid-mediated remodeling of the mammalian Na + /H + exchanger NHA2.
Nat.Struct.Mol.Biol., 29, 2022
|
|
7SHT
 
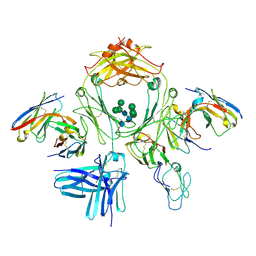 | |
7SHY
 
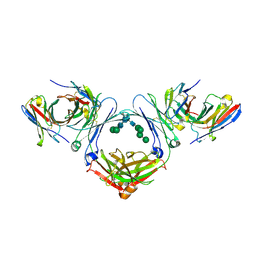 | | IgE-Fc in complex with omalizumab scFv | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, GLYCEROL, IgE Fc, ... | | Authors: | Pennington, L.F, Jardetzky, T.J, Kleinboelting, S. | | Deposit date: | 2021-10-11 | | Release date: | 2021-12-15 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (3 Å) | | Cite: | Directed evolution of and structural insights into antibody-mediated disruption of a stable receptor-ligand complex.
Nat Commun, 12, 2021
|
|
7SI0
 
 | | IgE-Fc in complex with 813 | | Descriptor: | 813 Variable fragment Heavy chain, 813 Variable fragment Light chain, IgE Fc, ... | | Authors: | Pennington, L.F, Jardetzky, T.J, Kleinboelting, S. | | Deposit date: | 2021-10-12 | | Release date: | 2021-12-15 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (3 Å) | | Cite: | Directed evolution of and structural insights into antibody-mediated disruption of a stable receptor-ligand complex.
Nat Commun, 12, 2021
|
|
7SHU
 
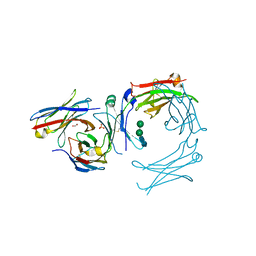 | | IgE-Fc in complex with omalizumab variant C02 | | Descriptor: | 1,2-ETHANEDIOL, 2-acetamido-2-deoxy-beta-D-glucopyranose, Immunoglobulin heavy constant epsilon, ... | | Authors: | Pennington, L.F, Jardetzky, T.J, Kleinboelting, S. | | Deposit date: | 2021-10-11 | | Release date: | 2021-12-15 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (2.75 Å) | | Cite: | Directed evolution of and structural insights into antibody-mediated disruption of a stable receptor-ligand complex.
Nat Commun, 12, 2021
|
|
7SHZ
 
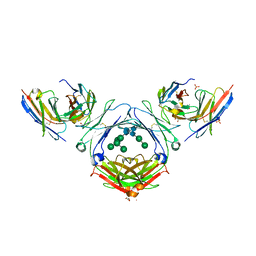 | | IgE-Fc in complex with HAE | | Descriptor: | 1,2-ETHANEDIOL, 2-acetamido-2-deoxy-beta-D-glucopyranose, ACETATE ION, ... | | Authors: | Pennington, L.F, Jardetzky, T.J, Kleinboelting, S. | | Deposit date: | 2021-10-11 | | Release date: | 2021-12-22 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (3 Å) | | Cite: | Directed evolution of and structural insights into antibody-mediated disruption of a stable receptor-ligand complex.
Nat Commun, 12, 2021
|
|
8HZT
 
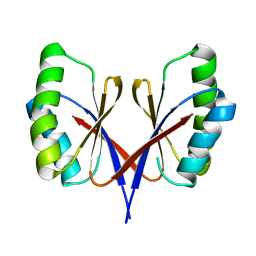 | |
8HZQ
 
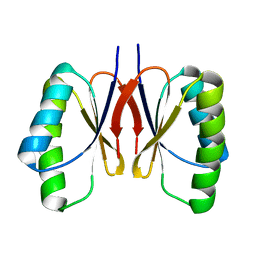 | | Bacillus subtilis SepF protein assembly (wild type) | | Descriptor: | Cell division protein SepF | | Authors: | Liu, W. | | Deposit date: | 2023-01-09 | | Release date: | 2024-02-14 | | Last modified: | 2024-05-15 | | Method: | SOLID-STATE NMR | | Cite: | Molecular basis for curvature formation in SepF polymerization.
Proc.Natl.Acad.Sci.USA, 121, 2024
|
|
5WJ6
 
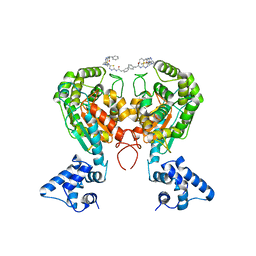 | | Crystal structure of glutaminase C in complex with inhibitor 2-phenyl-N-{5-[4-({5-[(phenylacetyl)amino]-1,3,4-thiadiazol-2-yl}amino)piperidin-1-yl]-1,3,4-thiadiazol-2-yl}acetamide (UPGL-00004) | | Descriptor: | 2-phenyl-N-{5-[4-({5-[(phenylacetyl)amino]-1,3,4-thiadiazol-2-yl}amino)piperidin-1-yl]-1,3,4-thiadiazol-2-yl}acetamide, Glutaminase kidney isoform, mitochondrial | | Authors: | Huang, Q, Cerione, R.A. | | Deposit date: | 2017-07-21 | | Release date: | 2018-01-10 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (2.445 Å) | | Cite: | Characterization of the interactions of potent allosteric inhibitors with glutaminase C, a key enzyme in cancer cell glutamine metabolism.
J. Biol. Chem., 293, 2018
|
|
5WXL
 
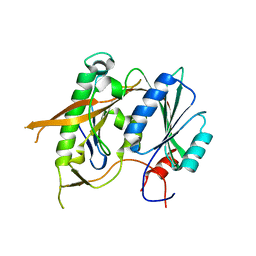 | | Crystal structure of the Rrs1 and Rpf2 complex | | Descriptor: | Regulator of ribosome biosynthesis, Ribosome biogenesis protein RPF2 | | Authors: | Ye, K, Zheng, S. | | Deposit date: | 2017-01-07 | | Release date: | 2017-01-25 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Molecular architecture of the 90S small subunit pre-ribosome
Elife, 6, 2017
|
|
5WYJ
 
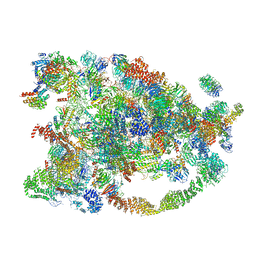 | | Cryo-EM structure of the 90S small subunit pre-ribosome (Dhr1-depleted, Enp1-TAP, state 1) | | Descriptor: | 13 kDa ribonucleoprotein-associated protein, 18S ribosomal RNA, 40S ribosomal protein S1-A, ... | | Authors: | Ye, K, Zhu, X, Sun, Q. | | Deposit date: | 2017-01-13 | | Release date: | 2017-03-29 | | Last modified: | 2019-10-09 | | Method: | ELECTRON MICROSCOPY (8.7 Å) | | Cite: | Molecular architecture of the 90S small subunit pre-ribosome.
Elife, 6, 2017
|
|
5WWN
 
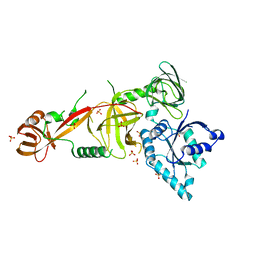 | | Crystal structure of Tsr1 | | Descriptor: | Ribosome biogenesis protein TSR1, SULFATE ION | | Authors: | Ye, K, Wang, B. | | Deposit date: | 2017-01-03 | | Release date: | 2017-06-28 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (2.805 Å) | | Cite: | Molecular architecture of the 90S small subunit pre-ribosome
Elife, 6, 2017
|
|
5WXM
 
 | | Crystal structure of the Imp3 and Mpp10 complex | | Descriptor: | SULFATE ION, U3 small nucleolar RNA-associated protein MPP10, U3 small nucleolar ribonucleoprotein protein IMP3 | | Authors: | Ye, K, Zheng, S. | | Deposit date: | 2017-01-07 | | Release date: | 2017-06-28 | | Method: | X-RAY DIFFRACTION (2.304 Å) | | Cite: | Molecular architecture of the 90S small subunit pre-ribosome
Elife, 6, 2017
|
|
5WWM
 
 | | Crystal structure of the TPR domain of Rrp5 | | Descriptor: | SULFATE ION, rRNA biogenesis protein RRP5 | | Authors: | Ye, K, Chen, X. | | Deposit date: | 2017-01-03 | | Release date: | 2017-01-25 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (2.809 Å) | | Cite: | Correction: Molecular architecture of the 90S small subunit pre-ribosome
Elife, 6, 2017
|
|
