1MQ9
 
 | | Crystal structure of high affinity alphaL I domain with ligand mimetic crystal contact | | Descriptor: | Integrin alpha-L, MANGANESE (II) ION | | Authors: | Shimaoka, M, Xiao, T, Liu, J.-H, Yang, Y, Dong, Y, Jun, C.-D, McCormack, A, Zhang, R, Joachimiak, A, Takagi, J, Wang, J.-H, Springer, T.A. | | Deposit date: | 2002-09-15 | | Release date: | 2003-01-14 | | Last modified: | 2021-10-27 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Structures of the aL I domain and its complex with ICAM-1 reveal a shape-shifting pathway for integrin regulation
Cell(Cambridge,Mass.), 112, 2003
|
|
5FU5
 
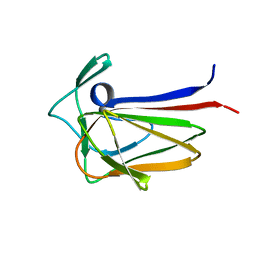 | |
3TUC
 
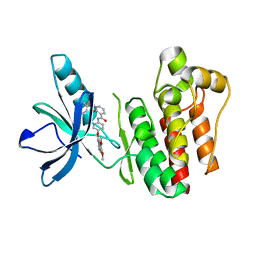 | | Crystal structure of SYK kinase domain with 1-benzyl-N-(5-(6,7-dimethoxyquinolin-4-yloxy)pyridin-2-yl)-2-oxo-1,2-dihydropyridine-3-carboxamide | | Descriptor: | 1-benzyl-N-{5-[(6,7-dimethoxyquinolin-4-yl)oxy]pyridin-2-yl}-2-oxo-1,2-dihydropyridine-3-carboxamide, Tyrosine-protein kinase SYK | | Authors: | Lovering, F, McDonald, J, Whitlock, G, Glossop, P, Phillips, C, Sabnis, Y, Ryan, M, Fitz, L, Lee, J, Chang, J.S, Han, S, Kurumbail, R, Thorarenson, A. | | Deposit date: | 2011-09-16 | | Release date: | 2012-08-29 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Identification of Type-II Inhibitors Using Kinase Structures.
Chem.Biol.Drug Des., 80, 2012
|
|
3U9A
 
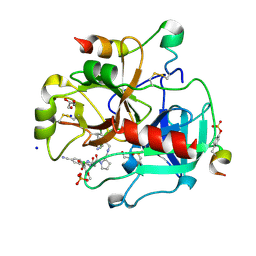 | | Human Thrombin In Complex With MI330 | | Descriptor: | (2S)-N-[[2-(aminomethyl)-5-chloranyl-phenyl]methyl]-1-[(2S)-2-[(3-chloranyl-4-methoxy-phenyl)sulfonylamino]-4-[(4-cyanophenyl)methylamino]-4-oxidanylidene-butanoyl]pyrrolidine-2-carboxamide, 2-acetamido-2-deoxy-beta-D-glucopyranose, GLYCEROL, ... | | Authors: | Biela, A, Heine, A, Klebe, G. | | Deposit date: | 2011-10-18 | | Release date: | 2012-10-24 | | Last modified: | 2023-12-06 | | Method: | X-RAY DIFFRACTION (1.58 Å) | | Cite: | Thrombin Inhibition
To be Published
|
|
3TW2
 
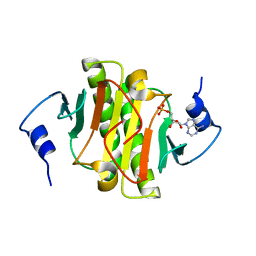 | | High resolution structure of human histidine triad nucleotide-binding protein 1 (hHINT1)/AMP complex in a monoclinic space group | | Descriptor: | ADENOSINE MONOPHOSPHATE, Histidine triad nucleotide-binding protein 1 | | Authors: | Dolot, R.M, Wlodarczyk, A, Ozga, M, Krakowiak, A, Nawrot, B. | | Deposit date: | 2011-09-21 | | Release date: | 2011-11-02 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (1.38 Å) | | Cite: | A new crystal form of human histidine triad nucleotide-binding protein 1 (hHINT1) in complex with adenosine 5'-monophosphate at 1.38 A resolution.
Acta Crystallogr.,Sect.F, 68, 2012
|
|
4MZP
 
 | | MazF from S. aureus crystal form III, C2221, 2.7 A | | Descriptor: | MazF mRNA interferase | | Authors: | Zorzini, V, Loris, R, van Nuland, N.A.J, Cheung, A. | | Deposit date: | 2013-09-30 | | Release date: | 2014-05-21 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (2.702 Å) | | Cite: | Structural and biophysical characterization of Staphylococcus aureus SaMazF shows conservation of functional dynamics.
Nucleic Acids Res., 42, 2014
|
|
1MOS
 
 | | ISOMERASE DOMAIN OF GLUCOSAMINE 6-PHOSPHATE SYNTHASE COMPLEXED WITH 2-AMINO-2-DEOXYGLUCITOL 6-PHOSPHATE | | Descriptor: | 2-(N-MORPHOLINO)-ETHANESULFONIC ACID, 2-DEOXY-2-AMINO GLUCITOL-6-PHOSPHATE, GLUCOSAMINE 6-PHOSPHATE SYNTHASE, ... | | Authors: | Teplyakov, A, Obmolova, G, Badet-Denisot, M.A, Badet, B. | | Deposit date: | 1998-07-15 | | Release date: | 1999-07-29 | | Last modified: | 2024-05-22 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | The mechanism of sugar phosphate isomerization by glucosamine 6-phosphate synthase.
Protein Sci., 8, 1999
|
|
1MDV
 
 | | KEY ROLE OF PHENYLALANINE 20 IN CYTOCHROME C3: STRUCTURE, STABILITY AND FUNCTION STUDIES | | Descriptor: | CYTOCHROME C3, PROTOPORPHYRIN IX CONTAINING FE | | Authors: | Dolla, A, Arnoux, P, Protasevich, I, Lobachov, V, Brugna, M, Guidici-Orticoni, M.T, Haser, R, Czjzek, M, Makarov, A, Brushi, M. | | Deposit date: | 1998-09-08 | | Release date: | 1999-05-04 | | Last modified: | 2023-08-09 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Key role of phenylalanine 20 in cytochrome c3: structure, stability, and function studies.
Biochemistry, 38, 1999
|
|
5FGK
 
 | | CDK8-CYCC IN COMPLEX WITH 8-[3-(3-Amino-1H-indazol-6-yl)-5-chloro- pyridine-4-yl]-2,8-diaza-spiro[4.5]decan-1-one | | Descriptor: | 1,2-ETHANEDIOL, 8-[3-(3-azanyl-2~{H}-indazol-6-yl)-5-chloranyl-pyridin-4-yl]-2,8-diazaspiro[4.5]decan-1-one, Cyclin-C, ... | | Authors: | Musil, D, Blagg, J, Mallinger, A. | | Deposit date: | 2015-12-20 | | Release date: | 2016-02-03 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (2.36 Å) | | Cite: | Discovery of Potent, Selective, and Orally Bioavailable Small-Molecule Modulators of the Mediator Complex-Associated Kinases CDK8 and CDK19.
J.Med.Chem., 59, 2016
|
|
1MBJ
 
 | | MOUSE C-MYB DNA-BINDING DOMAIN REPEAT 3 | | Descriptor: | MYB PROTO-ONCOGENE PROTEIN | | Authors: | Ogata, K, Morikawa, S, Nakamura, H, Hojo, H, Yoshimura, S, Zhang, R, Aimoto, S, Ametani, Y, Hirata, Z, Sarai, A, Ishii, S, Nishimura, Y. | | Deposit date: | 1995-05-19 | | Release date: | 1995-07-31 | | Last modified: | 2024-06-05 | | Method: | SOLUTION NMR | | Cite: | Comparison of the free and DNA-complexed forms of the DNA-binding domain from c-Myb.
Nat.Struct.Biol., 2, 1995
|
|
1MBF
 
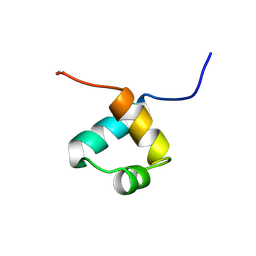 | | MOUSE C-MYB DNA-BINDING DOMAIN REPEAT 1 | | Descriptor: | MYB PROTO-ONCOGENE PROTEIN | | Authors: | Ogata, K, Morikawa, S, Nakamura, H, Hojo, H, Yoshimura, S, Zhang, R, Aimoto, S, Ametani, Y, Hirata, Z, Sarai, A, Ishii, S, Nishimura, Y. | | Deposit date: | 1995-05-19 | | Release date: | 1995-07-31 | | Last modified: | 2024-06-05 | | Method: | SOLUTION NMR | | Cite: | Comparison of the free and DNA-complexed forms of the DNA-binding domain from c-Myb.
Nat.Struct.Biol., 2, 1995
|
|
4MUC
 
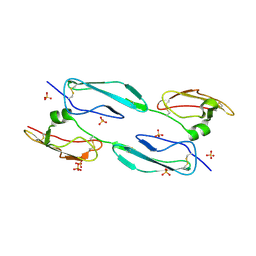 | | The 4th and 5th C-terminal domains of Factor H related protein 1 | | Descriptor: | Complement factor H-related protein 1, SULFATE ION | | Authors: | Bhattacharjee, A, Goldman, A, Kolodziejczyk, R, Jokiranta, T.S. | | Deposit date: | 2013-09-21 | | Release date: | 2015-02-18 | | Last modified: | 2015-05-06 | | Method: | X-RAY DIFFRACTION (2.897 Å) | | Cite: | The Major Autoantibody Epitope on Factor H in Atypical Hemolytic Uremic Syndrome Is Structurally Different from Its Homologous Site in Factor H-related Protein 1, Supporting a Novel Model for Induction of Autoimmunity in This Disease.
J.Biol.Chem., 290, 2015
|
|
1MBK
 
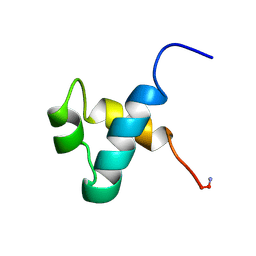 | | MOUSE C-MYB DNA-BINDING DOMAIN REPEAT 3 | | Descriptor: | MYB PROTO-ONCOGENE PROTEIN | | Authors: | Ogata, K, Morikawa, S, Nakamura, H, Hojo, H, Yoshimura, S, Zhang, R, Aimoto, S, Ametani, Y, Hirata, Z, Sarai, A, Ishii, S, Nishimura, Y. | | Deposit date: | 1995-05-19 | | Release date: | 1995-07-31 | | Last modified: | 2024-06-05 | | Method: | SOLUTION NMR | | Cite: | Comparison of the free and DNA-complexed forms of the DNA-binding domain from c-Myb.
Nat.Struct.Biol., 2, 1995
|
|
3U27
 
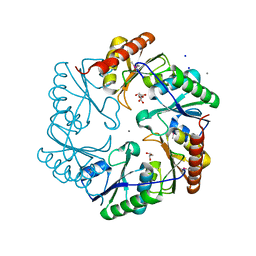 | | Crystal structure of ethanolamine utilization protein EutL from Leptotrichia buccalis C-1013-b | | Descriptor: | CALCIUM ION, GLYCEROL, Microcompartments protein, ... | | Authors: | Wu, R, Gu, M, Kerfeld, C.A, Salmeen, A, Joachimiak, A, Midwest Center for Structural Genomics (MCSG) | | Deposit date: | 2011-10-01 | | Release date: | 2012-02-08 | | Last modified: | 2020-01-29 | | Method: | X-RAY DIFFRACTION (1.852 Å) | | Cite: | Crystal structure of ethanolamine utilization protein EutL from Leptotrichia buccalis C-1013-b
To be Published
|
|
1MQR
 
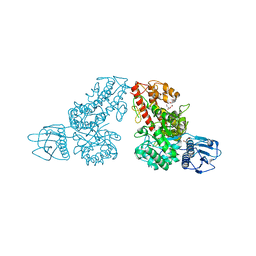 | | THE CRYSTAL STRUCTURE OF ALPHA-D-GLUCURONIDASE (E386Q) FROM BACILLUS STEAROTHERMOPHILUS T-6 | | Descriptor: | ALPHA-D-GLUCURONIDASE, GLYCEROL | | Authors: | Golan, G, Shallom, D, Teplitsky, A, Zaide, G, Shulami, S, Baasov, T, Stojanoff, V, Thompson, A, Shoham, Y, Shoham, G. | | Deposit date: | 2002-09-17 | | Release date: | 2003-09-17 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Crystal Structures of Geobacillus stearothermophilus {alpha}-Glucuronidase Complexed with Its Substrate and Products: MECHANISTIC IMPLICATIONS.
J.Biol.Chem., 279, 2004
|
|
1MYW
 
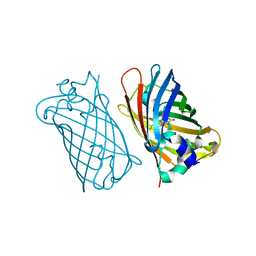 | | CRYSTAL STRUCTURE OF A YELLOW FLUORESCENT PROTEIN WITH IMPROVED MATURATION AND REDUCED ENVIRONMENTAL SENSITIVITY | | Descriptor: | Green fluorescent protein | | Authors: | Rekas, A, Alattia, J.R, Nagai, T, Miyawaki, A, Ikura, M. | | Deposit date: | 2002-10-04 | | Release date: | 2003-01-14 | | Last modified: | 2021-10-27 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Crystal Structure of Venus, a Yellow Fluorescent
Protein with Improved Maturation and
Reduced Environmental Sensitivity
J.Biol.Chem., 277, 2002
|
|
1MW5
 
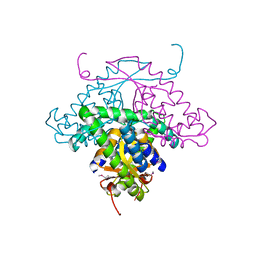 | | Structure of HI1480 from Haemophilus influenzae | | Descriptor: | HYPOTHETICAL PROTEIN HI1480 | | Authors: | Lim, K, Sarikaya, E, Howard, A, Galkin, A, Herzberg, O, Structure 2 Function Project (S2F) | | Deposit date: | 2002-09-27 | | Release date: | 2003-11-18 | | Last modified: | 2017-10-11 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Novel structure and nucleotide binding properties of HI1480 from Haemophilus influenzae: a protein with no known sequence homologues
PROTEINS: STRUCT.,FUNCT.,GENET., 56, 2004
|
|
1MQP
 
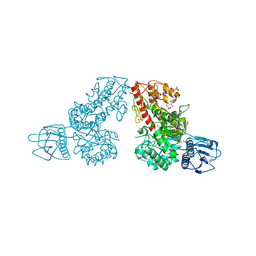 | | THE CRYSTAL STRUCTURE OF ALPHA-D-GLUCURONIDASE FROM BACILLUS STEAROTHERMOPHILUS T-6 | | Descriptor: | GLYCEROL, alpha-D-glucuronidase | | Authors: | Golan, G, Shallom, D, Teplitsky, A, Zaide, G, Shulami, S, Baasov, T, Stojanoff, V, Thompson, A, Shoham, Y, Shoham, G. | | Deposit date: | 2002-09-17 | | Release date: | 2003-09-23 | | Last modified: | 2011-07-13 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Crystal structures of Geobacillus stearothermophilus alpha-glucuronidase complexed with its substrate and products: mechanistic implications.
J.Biol.Chem., 279, 2004
|
|
5FU3
 
 | | The complexity of the Ruminococcus flavefaciens cellulosome reflects an expansion in glycan recognition | | Descriptor: | CBM74-RFGH5, SODIUM ION, beta-D-glucopyranose-(1-4)-beta-D-glucopyranose-(1-4)-beta-D-glucopyranose | | Authors: | Basle, A, Luis, A.S, Venditto, I, Gilbert, H.J. | | Deposit date: | 2016-01-20 | | Release date: | 2016-06-22 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (1.61 Å) | | Cite: | Complexity of the Ruminococcus Flavefaciens Cellulosome Reflects an Expansion in Glycan Recognition.
Proc.Natl.Acad.Sci.USA, 113, 2016
|
|
1MQQ
 
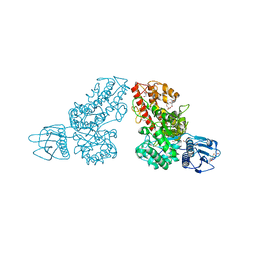 | | THE CRYSTAL STRUCTURE OF ALPHA-D-GLUCURONIDASE FROM BACILLUS STEAROTHERMOPHILUS T-1 COMPLEXED WITH GLUCURONIC ACID | | Descriptor: | ALPHA-D-GLUCURONIDASE, GLYCEROL, alpha-D-glucopyranuronic acid | | Authors: | Golan, G, Shallom, D, Teplitsky, A, Zaide, G, Shulami, S, Baasov, T, Stojanoff, V, Thompson, A, Shoham, Y, Shoham, G. | | Deposit date: | 2002-09-17 | | Release date: | 2003-09-17 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (1.65 Å) | | Cite: | Crystal structures of Geobacillus stearothermophilus alpha-glucuronidase complexed with its substrate and products: mechanistic implications.
J.Biol.Chem., 279, 2004
|
|
2QQV
 
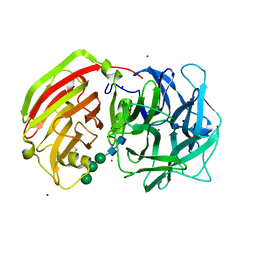 | | Crystal structure of a cell-wall invertase (E203A) from Arabidopsis thaliana in complex with sucrose | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Beta-fructofuranosidase, ... | | Authors: | Lammens, W, Le Roy, K, Van Laere, A, Rabijns, A, Van den Ende, W. | | Deposit date: | 2007-07-27 | | Release date: | 2008-04-22 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (3.01 Å) | | Cite: | Crystal structures of Arabidopsis thaliana cell-wall invertase mutants in complex with sucrose.
J.Mol.Biol., 377, 2008
|
|
5FBU
 
 | | Crystal structure of rifampin phosphotransferase RPH-Lm from Listeria monocytogenes in complex with rifampin-phosphate | | Descriptor: | (4S)-2-METHYL-2,4-PENTANEDIOL, CHLORIDE ION, Phosphoenolpyruvate synthase, ... | | Authors: | Stogios, P.J, Wawrzak, Z, Skarina, T, Yim, V, Savchenko, A, Anderson, W.F, Center for Structural Genomics of Infectious Diseases (CSGID) | | Deposit date: | 2015-12-14 | | Release date: | 2015-12-30 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (2.85 Å) | | Cite: | Rifampin phosphotransferase is an unusual antibiotic resistance kinase.
Nat Commun, 7, 2016
|
|
1N3B
 
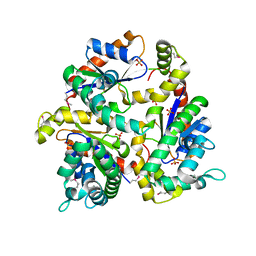 | | Crystal Structure of Dephosphocoenzyme A kinase from Escherichia coli | | Descriptor: | Dephospho-CoA kinase, SULFATE ION | | Authors: | O'Toole, N, Barbosa, J.A.R.G, Li, Y, Hung, L.-W, Matte, A, Cygler, M, Montreal-Kingston Bacterial Structural Genomics Initiative (BSGI) | | Deposit date: | 2002-10-25 | | Release date: | 2003-01-28 | | Last modified: | 2017-02-01 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Crystal Structure of a Trimeric Form of Dephosphocoenzyme A Kinase from Escherichia coli
Protein Sci., 12, 2003
|
|
1N3R
 
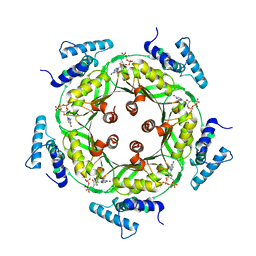 | | Biosynthesis of pteridins. Reaction mechanism of GTP cyclohydrolase I | | Descriptor: | GTP cyclohydrolase I, GUANOSINE-5'-TRIPHOSPHATE | | Authors: | Rebelo, J, Auerbach, G, Bader, G, Bracher, A, Nar, H, Hoesl, C, Schramek, N, Kaiser, J, Bacher, A, Huber, R, Fischer, M. | | Deposit date: | 2002-10-29 | | Release date: | 2003-10-14 | | Last modified: | 2024-05-29 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Biosynthesis of Pteridines. Reaction Mechanism of GTP Cyclohydrolase I
J.MOL.BIOL., 326, 2003
|
|
8C77
 
 | | Human cathepsin L after reaction with the thiocarbazate inhibitor CID 16725315 | | Descriptor: | 1,2-ETHANEDIOL, ACETATE ION, Cathepsin L, ... | | Authors: | Falke, S, Lieske, J, Guenther, S, Reinke, P.Y.A, Ewert, W, Loboda, J, Karnicar, K, Usenik, A, Lindic, N, Sekirnik, A, Chapman, H.N, Hinrichs, W, Turk, D, Meents, A. | | Deposit date: | 2023-01-12 | | Release date: | 2023-01-25 | | Last modified: | 2024-05-22 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Structural Elucidation and Antiviral Activity of Covalent Cathepsin L Inhibitors.
J.Med.Chem., 67, 2024
|
|
