6BP7
 
 | | Recombinant major vault protein [Rattus norvegicus] structure in solution: conformation 2 | | Descriptor: | Major vault protein | | Authors: | Ding, K, Zhang, X, Mrazek, J, Kickhoefer, V.A, Lai, M, Ng, H.L, Yang, O.O, Rome, L.H, Zhou, Z.H. | | Deposit date: | 2017-11-22 | | Release date: | 2018-04-04 | | Last modified: | 2024-03-13 | | Method: | ELECTRON MICROSCOPY (4.9 Å) | | Cite: | Solution Structures of Engineered Vault Particles.
Structure, 26, 2018
|
|
4R2A
 
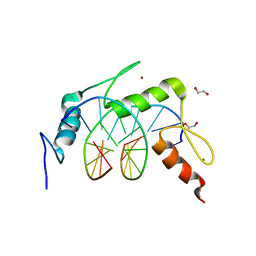 | | Egr1/Zif268 zinc fingers in complex with methylated DNA | | Descriptor: | 1,2-ETHANEDIOL, DNA (5'-D(*AP*GP*CP*GP*TP*GP*GP*GP*(5CM)P*GP*T)-3'), DNA (5'-D(*TP*AP*(5CM)P*GP*CP*CP*CP*AP*CP*GP*C)-3'), ... | | Authors: | Hashimoto, H, Olanrewaju, Y.O, Zheng, Y, Wilson, G.G, Zhang, X, Cheng, X. | | Deposit date: | 2014-08-11 | | Release date: | 2014-10-08 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (1.591 Å) | | Cite: | Wilms tumor protein recognizes 5-carboxylcytosine within a specific DNA sequence.
Genes Dev., 28, 2014
|
|
4R2D
 
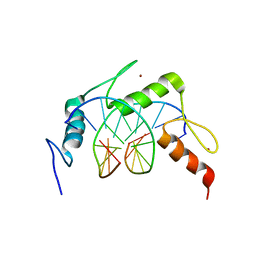 | | Egr1/Zif268 zinc fingers in complex with formylated DNA | | Descriptor: | DNA (5'-D(*AP*GP*CP*GP*TP*GP*GP*GP*(5FC)P*GP*T)-3'), DNA (5'-D(*TP*AP*(5FC)P*GP*CP*CP*CP*AP*CP*GP*C)-3'), Early growth response protein 1, ... | | Authors: | Hashimoto, H, Olanrewaju, Y.O, Zheng, Y, Wilson, G.G, Zhang, X, Cheng, X. | | Deposit date: | 2014-08-11 | | Release date: | 2014-10-08 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (2.088 Å) | | Cite: | Wilms tumor protein recognizes 5-carboxylcytosine within a specific DNA sequence.
Genes Dev., 28, 2014
|
|
4R2P
 
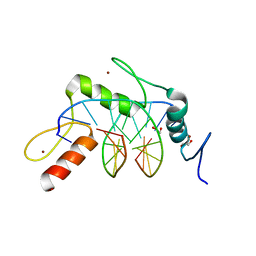 | | Wilms Tumor Protein (WT1) zinc fingers in complex with hydroxymethylated DNA | | Descriptor: | 1,2-ETHANEDIOL, DNA (5'-D(*AP*GP*CP*GP*TP*GP*GP*GP*(5HC)P*GP*T)-3'), DNA (5'-D(*TP*AP*(5HC)P*GP*CP*CP*CP*AP*CP*GP*C)-3'), ... | | Authors: | Hashimoto, H, Olanrewaju, Y.O, Zheng, Y, Wilson, G.G, Zhang, X, Cheng, X. | | Deposit date: | 2014-08-12 | | Release date: | 2014-10-08 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (1.788 Å) | | Cite: | Wilms tumor protein recognizes 5-carboxylcytosine within a specific DNA sequence.
Genes Dev., 28, 2014
|
|
4R2S
 
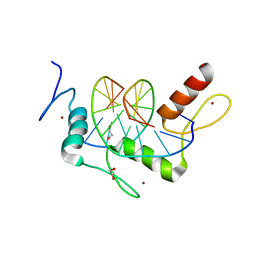 | | Wilms Tumor Protein (WT1) Q369P zinc fingers in complex with methylated DNA | | Descriptor: | 1,2-ETHANEDIOL, DNA (5'-D(*AP*GP*CP*GP*TP*GP*GP*GP*(5CM)P*GP*T)-3'), DNA (5'-D(*TP*AP*(5CM)P*GP*CP*CP*CP*AP*CP*GP*C)-3'), ... | | Authors: | Hashimoto, H, Olanrewaju, Y.O, Zheng, Y, Wilson, G.G, Zhang, X, Cheng, X. | | Deposit date: | 2014-08-12 | | Release date: | 2014-10-08 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (2.489 Å) | | Cite: | Wilms tumor protein recognizes 5-carboxylcytosine within a specific DNA sequence.
Genes Dev., 28, 2014
|
|
9L1N
 
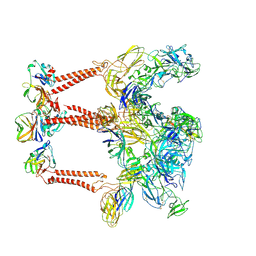 | | Structure of Western equine encephalitis virus 71V1658 strain VLP in complex with human PCDH10 EC1 | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, Capsid glycoprotein, E1 glycoprotein, ... | | Authors: | Cao, D, Ma, B, Cao, Z, Zhang, X, Xiang, Y. | | Deposit date: | 2024-12-15 | | Release date: | 2025-06-04 | | Method: | ELECTRON MICROSCOPY (3.3 Å) | | Cite: | Structural basis for the recognition of two different types of receptors by Western equine encephalitis virus.
Cell Rep, 44, 2025
|
|
8QOT
 
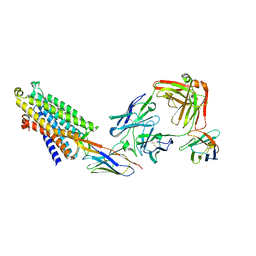 | | Structure of the mu opioid receptor bound to the antagonist nanobody NbE | | Descriptor: | Anti-Fab Nanobody, Mu-type opioid receptor, NabFab HC, ... | | Authors: | Yu, J, Kumar, A, Zhang, X, Martin, C, Raia, P, Manglik, A, Ballet, S, Boland, A, Stoeber, M. | | Deposit date: | 2023-09-29 | | Release date: | 2023-12-27 | | Last modified: | 2025-01-29 | | Method: | ELECTRON MICROSCOPY (3.2 Å) | | Cite: | Structural basis of mu-opioid receptor targeting by a nanobody antagonist.
Nat Commun, 15, 2024
|
|
4TOR
 
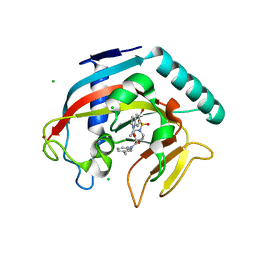 | | Crystal structure of Tankyrase 1 with IWR-8 | | Descriptor: | 1-[(1-acetyl-5-bromo-1H-indol-6-yl)sulfonyl]-N-ethyl-N-(3-methylphenyl)piperidine-4-carboxamide, CHLORIDE ION, Tankyrase-1, ... | | Authors: | Chen, H, Zhang, X, Lum, L, Chen, C. | | Deposit date: | 2014-06-06 | | Release date: | 2015-05-20 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (1.501 Å) | | Cite: | Disruption of Wnt/ beta-Catenin Signaling and Telomeric Shortening Are Inextricable Consequences of Tankyrase Inhibition in Human Cells.
Mol.Cell.Biol., 35, 2015
|
|
3EU9
 
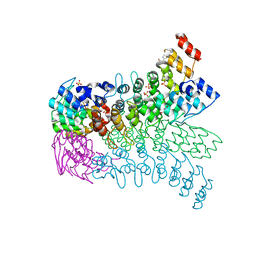 | | The ankyrin repeat domain of Huntingtin interacting protein 14 | | Descriptor: | GLYCEROL, HISTIDINE, Huntingtin-interacting protein 14, ... | | Authors: | Gao, T, Collins, R.E, Horton, J.R, Zhang, R, Zhang, X, Cheng, X. | | Deposit date: | 2008-10-09 | | Release date: | 2009-06-23 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (1.99 Å) | | Cite: | The ankyrin repeat domain of Huntingtin interacting protein 14 contains a surface aromatic cage, a potential site for methyl-lysine binding.
Proteins, 76, 2009
|
|
5ZG9
 
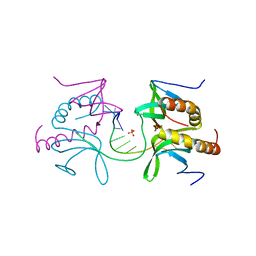 | | Crystal structure of MoSub1-ssDNA complex in phosphate buffer | | Descriptor: | DNA (5'-D(P*TP*TP*TP*TP*TP*TP*TP*TP*TP*TP*TP*TP*TP*TP*TP*TP*TP*TP*TP*G)-3'), MoSub1, PHOSPHATE ION | | Authors: | Zhao, Y, Huang, J, Liu, H, Yi, L, Wang, S, Zhang, X, Liu, J. | | Deposit date: | 2018-03-08 | | Release date: | 2019-03-27 | | Last modified: | 2024-03-27 | | Method: | X-RAY DIFFRACTION (2.04 Å) | | Cite: | The effect of phosphate ion on the ssDNA binding mode of MoSub1, a Sub1/PC4 homolog from rice blast fungus.
Proteins, 87, 2019
|
|
8QN4
 
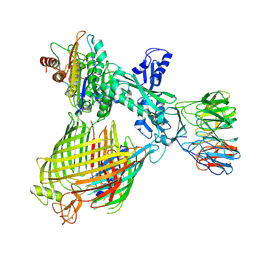 | | Structure of BAM-EspP complex in the non-closing EspP state | | Descriptor: | EspP epsilon, Outer membrane protein assembly factor BamA, Outer membrane protein assembly factor BamB, ... | | Authors: | Xie, T, Pang, J, Shen, C, Chang, S, Tang, X, Zhang, X, Dong, H, Zhou, R. | | Deposit date: | 2023-09-25 | | Release date: | 2024-10-02 | | Last modified: | 2024-10-23 | | Method: | ELECTRON MICROSCOPY (3.36 Å) | | Cite: | Dynamic topology-mediated maturation of beta-barrel proteins in BAM-catalyzed folding
To Be Published
|
|
5ZC3
 
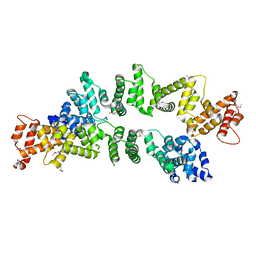 | | The Crystal Structure of PcRxLR12 | | Descriptor: | RxLR effector | | Authors: | Zhao, L, Zhang, X, Zhu, C. | | Deposit date: | 2018-02-14 | | Release date: | 2018-08-15 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (3.005 Å) | | Cite: | Crystal structure of the RxLR effector PcRxLR12 from Phytophthora capsici
Biochem. Biophys. Res. Commun., 503, 2018
|
|
4TOS
 
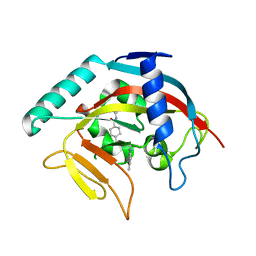 | | Crystal structure of Tankyrase 1 with 355 | | Descriptor: | Tankyrase-1, ZINC ION, trans-N-benzyl-4-({1-[(6-methyl-4-oxo-4H-pyrido[1,2-a]pyrimidin-2-yl)methyl]-2,4-dioxo-1,4-dihydroquinazolin-3(2H)-yl}methyl)cyclohexanecarboxamide | | Authors: | Chen, H, Zhang, X, Lum, l, Chen, C. | | Deposit date: | 2014-06-06 | | Release date: | 2015-05-20 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (1.802 Å) | | Cite: | Disruption of Wnt/ beta-Catenin Signaling and Telomeric Shortening Are Inextricable Consequences of Tankyrase Inhibition in Human Cells.
Mol.Cell.Biol., 35, 2015
|
|
8ZTC
 
 | | Crystal structure of Mps1-AMP complex | | Descriptor: | ADENOSINE MONOPHOSPHATE, Mitogen-activated protein kinase MPS1 | | Authors: | Kong, Z, Li, S, Zhang, X, Wang, D, Liu, J. | | Deposit date: | 2024-06-07 | | Release date: | 2025-04-23 | | Method: | X-RAY DIFFRACTION (2.26 Å) | | Cite: | Combinatorial Targeting of Common Docking and ATP Binding Sites on Mps1 MAPK for Management of Pathogenic Fungi.
J.Agric.Food Chem., 72, 2024
|
|
8ZMP
 
 | | Cryo-EM structure of the spike glycoprotein from Bat SARS-like coronavirus (Bat SL-CoV) WIV1 in locked state | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, LINOLEIC ACID, ... | | Authors: | Liu, C, Beck, F, Nagy, I, Bohn, S, Plitzko, J, Baumeister, W, Zhang, X, Zinzula, L. | | Deposit date: | 2024-05-23 | | Release date: | 2025-05-28 | | Method: | ELECTRON MICROSCOPY (3.38 Å) | | Cite: | Cryo-EM structure of the spike glycoprotein from Bat SARS-like coronavirus WIV1
To Be Published
|
|
4DFF
 
 | | The SAR development of dihydroimidazoisoquinoline derivatives as phosphodiesterase 10A inhibitors for the treatment of schizophrenia | | Descriptor: | 8,9-dimethoxy-1-(1,3-thiazol-5-yl)-5,6-dihydroimidazo[5,1-a]isoquinoline, MAGNESIUM ION, ZINC ION, ... | | Authors: | Ho, G.D, Seganish, W.M, Bercovici, A, Tulshian, D, Greenlee, W.J, Van Rijn, R, Hruza, A, Xiao, L, Rindgen, D, Mullins, D, Guzzi, M, Zhang, X, Bleichardt, C, Hodgson, R. | | Deposit date: | 2012-01-23 | | Release date: | 2012-03-14 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (2.11 Å) | | Cite: | The SAR development of dihydroimidazoisoquinoline derivatives as phosphodiesterase 10A inhibitors for the treatment of schizophrenia.
Bioorg.Med.Chem.Lett., 22, 2012
|
|
4DHI
 
 | | Structure of C. elegans OTUB1 bound to human UBC13 | | Descriptor: | Ubiquitin thioesterase otubain-like, Ubiquitin-conjugating enzyme E2 N | | Authors: | Wiener, R, Zhang, X, Wang, T, Wolberger, C. | | Deposit date: | 2012-01-27 | | Release date: | 2012-02-22 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | The mechanism of OTUB1-mediated inhibition of ubiquitination.
Nature, 483, 2012
|
|
4DHJ
 
 | | The structure of a ceOTUB1 ubiquitin aldehyde UBC13~Ub complex | | Descriptor: | Ubiquitin, Ubiquitin aldehyde, Ubiquitin thioesterase otubain-like, ... | | Authors: | Wiener, R, Zhang, X, Wang, T, Wolberger, C. | | Deposit date: | 2012-01-27 | | Release date: | 2012-02-22 | | Last modified: | 2025-03-26 | | Method: | X-RAY DIFFRACTION (2.35 Å) | | Cite: | The mechanism of OTUB1-mediated inhibition of ubiquitination.
Nature, 483, 2012
|
|
4DKT
 
 | | Crystal structure of human peptidylarginine deiminase 4 in complex with N-acetyl-L-threonyl-L-alpha-aspartyl-N5-[(1E)-2-fluoroethanimidoyl]-L-ornithinamide | | Descriptor: | 1,2-ETHANEDIOL, CALCIUM ION, Protein-arginine deiminase type-4, ... | | Authors: | Jones, J.E, Slack, J.L, Fang, P, Zhang, X, Subramanian, V, Causey, C.P, Coonrod, S.A, Guo, M, Thompson, P.R. | | Deposit date: | 2012-02-04 | | Release date: | 2012-02-29 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (2.98 Å) | | Cite: | Synthesis and Screening of a Haloacetamidine Containing Library To Identify PAD4 Selective Inhibitors.
Acs Chem.Biol., 7, 2012
|
|
4F5X
 
 | | Location of the dsRNA-dependent polymerase, VP1, in rotavirus particles | | Descriptor: | Intermediate capsid protein VP6, RNA-directed RNA polymerase, VP2 protein, ... | | Authors: | Estrozi, L.F, Settembre, E.C, Goret, G, McClain, B, Zhang, X, Chen, J.Z, Grigorieff, N, Harrison, S.C. | | Deposit date: | 2012-05-13 | | Release date: | 2012-10-24 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (5 Å) | | Cite: | Location of the dsRNA-Dependent Polymerase, VP1, in Rotavirus Particles.
J.Mol.Biol., 425, 2013
|
|
4DHZ
 
 | | The structure of h/ceOTUB1-ubiquitin aldehyde-UBC13~Ub | | Descriptor: | Ubiquitin, Ubiquitin aldehyde, Ubiquitin thioesterase otubain-like, ... | | Authors: | Wiener, R, Zhang, X, Wang, T, Wolberger, C. | | Deposit date: | 2012-01-30 | | Release date: | 2012-02-22 | | Last modified: | 2025-03-26 | | Method: | X-RAY DIFFRACTION (3.11 Å) | | Cite: | The mechanism of OTUB1-mediated inhibition of ubiquitination.
Nature, 483, 2012
|
|
5Y9P
 
 | | Staphylococcus aureus RNase HII | | Descriptor: | GLYCEROL, Ribonuclease HII | | Authors: | Hang, T, Wu, M, Zhang, X. | | Deposit date: | 2017-08-26 | | Release date: | 2018-08-01 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Structural insights into a novel functional dimer of Staphylococcus aureus RNase HII
Biochem. Biophys. Res. Commun., 503, 2018
|
|
2K9X
 
 | | Solution structure of Urm1 from Trypanosoma brucei | | Descriptor: | Uncharacterized protein | | Authors: | Zhang, W, Zhang, J, Xu, C, Wang, T, Zhang, X, Tu, X. | | Deposit date: | 2008-10-27 | | Release date: | 2009-03-10 | | Last modified: | 2024-05-29 | | Method: | SOLUTION NMR | | Cite: | Solution structure of Urm1 from Trypanosoma brucei
Proteins, 75, 2009
|
|
4HIN
 
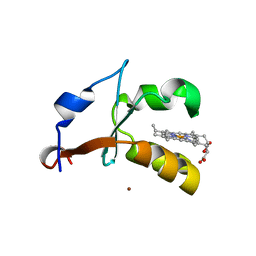 | | 2.4A Resolution Structure of Bovine Cytochrome b5 (S71L) | | Descriptor: | COPPER (II) ION, Cytochrome b5, PROTOPORPHYRIN IX CONTAINING FE | | Authors: | Lovell, S, Battaile, K.P, Parthasarathy, S, Sun, N, Terzyan, S, Zhang, X, Rivera, M, Kuczera, K, Benson, D.R. | | Deposit date: | 2012-10-11 | | Release date: | 2013-10-16 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | 2.4A Resolution Structure of Bovine Cytochrome b5 (S71L)
To be Published
|
|
4M8N
 
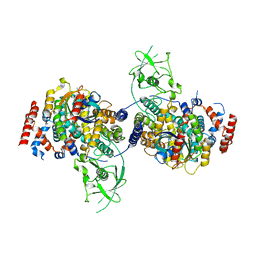 | | Crystal Structure of PlexinC1/Rap1B Complex | | Descriptor: | ALUMINUM FLUORIDE, GUANOSINE-5'-DIPHOSPHATE, MAGNESIUM ION, ... | | Authors: | Pascoe, H.G, Wang, Y, Brautigam, C.A, He, H, Zhang, X. | | Deposit date: | 2013-08-13 | | Release date: | 2013-10-09 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (3.294 Å) | | Cite: | Structural basis for activation and non-canonical catalysis of the Rap GTPase activating protein domain of plexin.
Elife, 2, 2013
|
|
