5XPW
 
 | | Structure of amphioxus IgVJ-C2 molecule | | Descriptor: | amphioxus IgVJ-C2 | | Authors: | Chen, R, Qi, J, Zhang, N, Zhang, L, Yao, S, Wu, Y, Jiang, B, Wang, Z, Yuan, H, Zhang, Q, Xia, C. | | Deposit date: | 2017-06-05 | | Release date: | 2018-04-18 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (2.003 Å) | | Cite: | Discovery and Analysis of Invertebrate IgVJ-C2 Structure from Amphioxus Provides Insight into the Evolution of the Ig Superfamily.
J. Immunol., 200, 2018
|
|
5XPV
 
 | | Structure of the V domain of amphioxus IgVJ-C2 | | Descriptor: | amphioxus IgVJ-C2 | | Authors: | Chen, R, Qi, J, Zhang, N, Zhang, L, Yao, S, Wu, Y, Jiang, B, Wang, Z, Yuan, H, Zhang, Q, Xia, C. | | Deposit date: | 2017-06-05 | | Release date: | 2018-04-18 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Discovery and Analysis of Invertebrate IgVJ-C2 Structure from Amphioxus Provides Insight into the Evolution of the Ig Superfamily.
J. Immunol., 200, 2018
|
|
3VW6
 
 | | Crystal structure of human apoptosis signal-regulating kinase 1 (ASK1) with imidazopyridine inhibitor | | Descriptor: | 4-tert-butyl-N-[6-(1H-imidazol-1-yl)imidazo[1,2-a]pyridin-2-yl]benzamide, Mitogen-activated protein kinase kinase kinase 5 | | Authors: | Terao, Y, Suzuki, H, Yoshikawa, M, Yashiro, H, Takekawa, S, Fujitani, Y, Okada, K, Inoue, Y, Yamamoto, Y, Nakagawa, H, Yao, S, Kawamoto, T, Uchikawa, O. | | Deposit date: | 2012-08-06 | | Release date: | 2012-10-31 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Design and biological evaluation of imidazo[1,2-a]pyridines as novel and potent ASK1 inhibitors.
Bioorg. Med. Chem. Lett., 22, 2012
|
|
1YXE
 
 | | Structure and inter-domain interactions of domain II from the blood stage malarial protein, apical membrane antigen 1 | | Descriptor: | apical membrane antigen 1 | | Authors: | Feng, Z.P, Keizer, D.W, Stevenson, R.A, Yao, S, Babon, J.J, Murphy, V.J, Anders, R.F, Norton, R.S. | | Deposit date: | 2005-02-21 | | Release date: | 2005-07-05 | | Last modified: | 2024-10-30 | | Method: | SOLUTION NMR | | Cite: | Structure and Inter-domain Interactions of Domain II from the Blood-stage Malarial Protein, Apical Membrane Antigen 1.
J.Mol.Biol., 350, 2005
|
|
6UEL
 
 | | CPS1 bound to allosteric inhibitor H3B-193 | | Descriptor: | Carbamoyl-phosphate synthase [ammonia], mitochondrial, N~1~-[(4-fluorophenyl)methyl]-N~1~-methyl-N~4~-(4-methyl-1,3-thiazol-2-yl)piperidine-1,4-dicarboxamide, ... | | Authors: | Larsen, N.A, Nguyen, T.V. | | Deposit date: | 2019-09-21 | | Release date: | 2020-03-18 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Small Molecule Inhibition of CPS1 Activity through an Allosteric Pocket.
Cell Chem Biol, 27, 2020
|
|
1DSJ
 
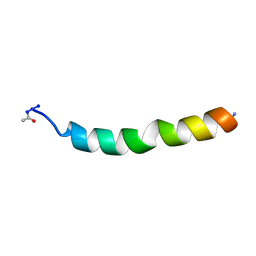 | | NMR SOLUTION STRUCTURE OF VPR50_75, 20 STRUCTURES | | Descriptor: | VPR PROTEIN | | Authors: | Yao, S, Torres, A.M, Azad, A.A, Macreadie, I.G, Norton, R.S. | | Deposit date: | 1997-10-23 | | Release date: | 1998-07-01 | | Last modified: | 2024-10-30 | | Method: | SOLUTION NMR | | Cite: | Helical structure of polypeptides from the C-terminal half of HIV-1 VPR.
Protein Pept.Lett., 5, 1998
|
|
7XF3
 
 | | The structure of HLA-B*1501/BM58-66AF9 | | Descriptor: | 9-mer peptide from Matrix protein 1, Beta-2-microglobulin, MHC class I antigen | | Authors: | Zhao, Y.Z, Xiao, W.L, Wu, Y.N, Fan, W.F, Yue, C, Zhang, Q.X, Zhang, D.N, Yuan, X.J, Yao, S.J, Liu, S, Li, M, Wang, P.Y, Zhang, H.J, Zhang, J, Zhao, M, Zheng, X.Q, Liu, W.J, Gao, G.F, Liu, W.L. | | Deposit date: | 2022-03-31 | | Release date: | 2023-02-08 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (1.91 Å) | | Cite: | Parallel T Cell Immunogenic Regions in Influenza B and A Viruses with Distinct Nuclear Export Signal Functions: The Balance between Viral Life Cycle and Immune Escape.
J Immunol., 210, 2023
|
|
6W2J
 
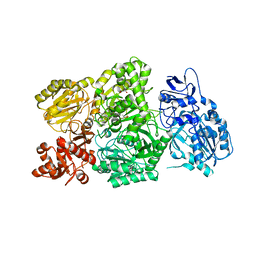 | | CPS1 bound to allosteric inhibitor H3B-374 | | Descriptor: | (2-fluoranyl-4-methoxy-phenyl)-[(3~{R},5~{R})-4-(2-fluoranyl-4-methoxy-phenyl)carbonyl-3,5-dimethyl-piperazin-1-yl]methanone, 1,2-ETHANEDIOL, Carbamoyl-phosphate synthase [ammonia], ... | | Authors: | Larsen, N.A, Nguyen, T.V. | | Deposit date: | 2020-03-05 | | Release date: | 2021-01-13 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (2.62 Å) | | Cite: | Discovery of 2,6-Dimethylpiperazines as Allosteric Inhibitors of CPS1.
Acs Med.Chem.Lett., 11, 2020
|
|
6JBT
 
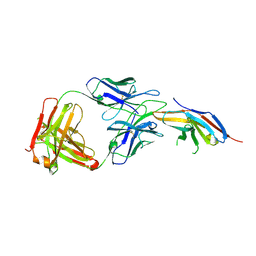 | | Complex structure of toripalimab-Fab and PD-1 | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-[alpha-L-fucopyranose-(1-6)]2-acetamido-2-deoxy-beta-D-glucopyranose, Heavy chain, ... | | Authors: | Guo, L, Tan, S, Chai, Y, Qi, J, Gao, G.F, Yan, J. | | Deposit date: | 2019-01-26 | | Release date: | 2019-06-19 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (2.47 Å) | | Cite: | Glycosylation-independent binding of monoclonal antibody toripalimab to FG loop of PD-1 for tumor immune checkpoint therapy.
Mabs, 11, 2019
|
|
5EOB
 
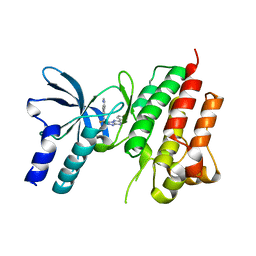 | | Crystal structure of CMET in complex with novel inhibitor | | Descriptor: | 6-[bis(fluoranyl)-[6-(4-fluorophenyl)-[1,2,4]triazolo[4,3-b][1,2,4]triazin-3-yl]methyl]quinoline, Hepatocyte growth factor receptor | | Authors: | Liu, Q, Chen, T, Xu, Y. | | Deposit date: | 2015-11-10 | | Release date: | 2016-10-19 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (1.75 Å) | | Cite: | Discovery of 6-(difluoro(6-(4-fluorophenyl)-[1,2,4]triazolo[4,3-b][1,2,4]triazin-3-yl)methyl)quinoline as a highly potent and selective c-Met inhibitor
Eur.J.Med.Chem., 116, 2016
|
|
5MAT
 
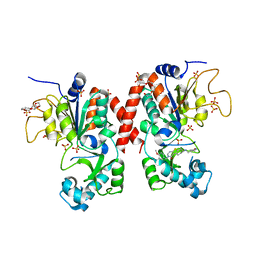 | | Structure of human Sirtuin 2 in complex with a selective thienopyrimidinone based inhibitor | | Descriptor: | (7~{R})-7-[(3,5-dimethyl-1,2-oxazol-4-yl)methylamino]-3-[(4-methoxynaphthalen-1-yl)methyl]-5,6,7,8-tetrahydro-[1]benzothiolo[2,3-d]pyrimidin-4-one, HEXAETHYLENE GLYCOL, NAD-dependent protein deacetylase sirtuin-2, ... | | Authors: | Moniot, S, Steegborn, C. | | Deposit date: | 2016-11-04 | | Release date: | 2017-02-08 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (2.069 Å) | | Cite: | Thienopyrimidinone Based Sirtuin-2 (SIRT2)-Selective Inhibitors Bind in the Ligand Induced Selectivity Pocket.
J. Med. Chem., 60, 2017
|
|
6CHZ
 
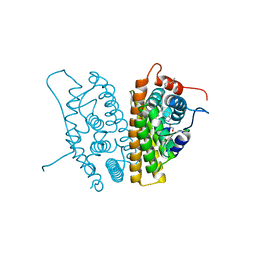 | | Estrogen Receptor Alpha Y537S bound to antagonist H3B-9224. | | Descriptor: | 1,2-ETHANEDIOL, 4-[(2-{4-[(1E)-1-(1H-indazol-5-yl)-2-phenylbut-1-en-1-yl]phenoxy}ethyl)amino]-N,N-dimethylbutanamide, Estrogen receptor | | Authors: | Larsen, N.A. | | Deposit date: | 2018-02-23 | | Release date: | 2018-03-21 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (1.68 Å) | | Cite: | Discovery of Selective Estrogen Receptor Covalent Antagonists for the Treatment of ER alphaWTand ER alphaMUTBreast Cancer.
Cancer Discov, 8, 2018
|
|
6CHW
 
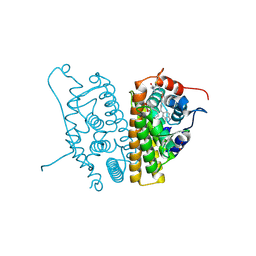 | | Estrogen Receptor Alpha Y537S covalently bound to antagonist H3B-5942. | | Descriptor: | 1,2-ETHANEDIOL, 4-[(2-{4-[(1E)-1-(1H-indazol-5-yl)-2-phenylbut-1-en-1-yl]phenoxy}ethyl)amino]-N,N-dimethylbutanamide, DIMETHYL SULFOXIDE, ... | | Authors: | Larsen, N.A. | | Deposit date: | 2018-02-23 | | Release date: | 2018-03-21 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (1.89 Å) | | Cite: | Discovery of Selective Estrogen Receptor Covalent Antagonists for the Treatment of ER alphaWTand ER alphaMUTBreast Cancer.
Cancer Discov, 8, 2018
|
|
6C2R
 
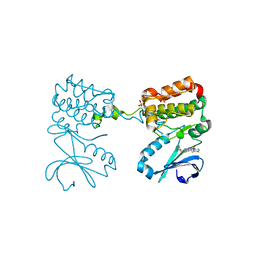 | | Aurora A ligand complex | | Descriptor: | (2R,4R)-1-[(3-chloro-2-fluorophenyl)methyl]-4-({3-fluoro-6-[(5-methyl-1H-pyrazol-3-yl)amino]pyridin-2-yl}methyl)-2-methylpiperidine-4-carboxylic acid, Aurora kinase A, SULFATE ION | | Authors: | Antonysamy, S, Pustilnik, A, Manglicmot, D, Froning, K, Weichert, K, Wasserman, S. | | Deposit date: | 2018-01-08 | | Release date: | 2019-01-23 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (1.96 Å) | | Cite: | Aurora A Kinase Inhibition Is Synthetic Lethal with Loss of theRB1Tumor Suppressor Gene.
Cancer Discov, 9, 2019
|
|
6C2T
 
 | | Aurora A ligand complex | | Descriptor: | (2S,4R)-1-[(3-chloro-2-fluorophenyl)methyl]-2-methyl-4-({3-[(1,3-thiazol-2-yl)amino]isoquinolin-1-yl}methyl)piperidine-4-carboxylic acid, Aurora kinase A, DIMETHYL SULFOXIDE, ... | | Authors: | Antonysamy, S, Pustilnik, A, Manglicmot, D, Froning, K, Weichert, K, Wasserman, S. | | Deposit date: | 2018-01-08 | | Release date: | 2019-01-23 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (1.73 Å) | | Cite: | Aurora A Kinase Inhibition Is Synthetic Lethal with Loss of theRB1Tumor Suppressor Gene.
Cancer Discov, 9, 2019
|
|
4DBN
 
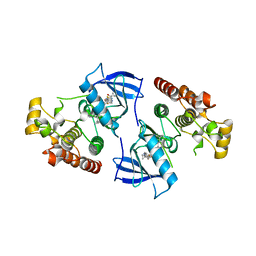 | | Crystal Structure of the Kinase domain of Human B-raf with a [1,3]thiazolo[5,4-b]pyridine derivative | | Descriptor: | 2-chloro-3-(1-cyanocyclopropyl)-N-[5-({2-[(cyclopropylcarbonyl)amino][1,3]thiazolo[5,4-b]pyridin-5-yl}oxy)-2-fluorophenyl]benzamide, Serine/threonine-protein kinase B-raf | | Authors: | Yano, J.K, Aertgeerts, K. | | Deposit date: | 2012-01-16 | | Release date: | 2012-04-11 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (3.15 Å) | | Cite: | Design and synthesis of novel DFG-out RAF/vascular endothelial growth factor receptor 2 (VEGFR2) inhibitors. 1. Exploration of [5,6]-fused bicyclic scaffolds.
J.Med.Chem., 55, 2012
|
|
3EMW
 
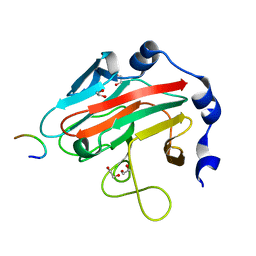 | | Crystal Structure of human splA/ryanodine receptor domain and SOCS box containing 2 (SPSB2) in complex with a 20-residue VASA peptide | | Descriptor: | 1,2-ETHANEDIOL, Peptide (VASA), SPRY domain-containing SOCS box protein 2 | | Authors: | Filippakopoulos, P, Sharpe, T, Keates, T, Murray, J.W, Savitsky, P, Roos, A.K, Pike, A.C.W, Von Delft, F, Arrowsmith, C.H, Edwards, A.M, Weigelt, J, Bountra, C, Knapp, S, Bullock, A, Structural Genomics Consortium (SGC) | | Deposit date: | 2008-09-25 | | Release date: | 2008-10-28 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Structural basis for Par-4 recognition by the SPRY domain- and SOCS box-containing proteins SPSB1, SPSB2, and SPSB4.
J.Mol.Biol., 401, 2010
|
|
3F2O
 
 | | Crystal Structure of human splA/ryanodine receptor domain and SOCS box containing 1 (SPSB1) in complex with a 20-residue VASA peptide | | Descriptor: | 20-mer peptide from ATP-dependent RNA helicase vasa, SPRY domain-containing SOCS box protein 1 | | Authors: | Filippakopoulos, P, Sharpe, T, Keates, T, Murray, J.W, Savitsky, P, Roos, A, Pike, A.C.W, Von Delft, F, Arrowsmith, C.H, Edwards, A.M, Weigelt, J, Bountra, C, Knapp, S, Bullock, A, Structural Genomics Consortium (SGC) | | Deposit date: | 2008-10-30 | | Release date: | 2008-12-09 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (2.05 Å) | | Cite: | Structural basis for Par-4 recognition by the SPRY domain- and SOCS box-containing proteins SPSB1, SPSB2, and SPSB4.
J.Mol.Biol., 401, 2010
|
|
4BD7
 
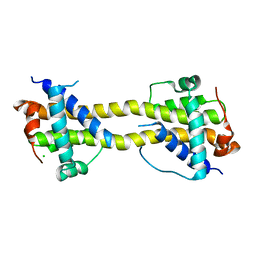 | | Bax domain swapped dimer induced by octylmaltoside | | Descriptor: | APOPTOSIS REGULATOR BAX, CHLORIDE ION, PRASEODYMIUM ION | | Authors: | Czabotar, P.E, Westphal, D, Adams, J.M, Colman, P.M. | | Deposit date: | 2012-10-05 | | Release date: | 2013-02-13 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (2.801 Å) | | Cite: | Bax Crystal Structures Reveal How Bh3 Domains Activate Bax and Nucleate its Oligomerization to Induce Apoptosis.
Cell(Cambridge,Mass.), 152, 2013
|
|
4BD2
 
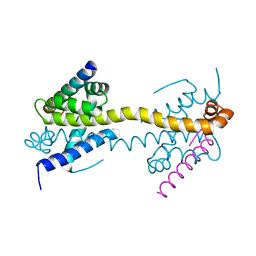 | | Bax domain swapped dimer in complex with BidBH3 | | Descriptor: | APOPTOSIS REGULATOR BAX, BH3-INTERACTING DOMAIN DEATH AGONIST | | Authors: | Czabotar, P.E, Westphal, D, Adams, J.M, Colman, P.M. | | Deposit date: | 2012-10-04 | | Release date: | 2013-02-13 | | Last modified: | 2024-05-01 | | Method: | X-RAY DIFFRACTION (2.206 Å) | | Cite: | Bax Crystal Structures Reveal How Bh3 Domains Activate Bax and Nucleate its Oligomerization to Induce Apoptosis.
Cell(Cambridge,Mass.), 152, 2013
|
|
4BD6
 
 | | Bax domain swapped dimer in complex with BaxBH3 | | Descriptor: | APOPTOSIS REGULATOR BAX | | Authors: | Czabotar, P.E, Westphal, D, Adams, J.M, Colman, P.M. | | Deposit date: | 2012-10-05 | | Release date: | 2013-02-13 | | Last modified: | 2024-05-01 | | Method: | X-RAY DIFFRACTION (2.494 Å) | | Cite: | Bax Crystal Structures Reveal How Bh3 Domains Activate Bax and Nucleate its Oligomerization to Induce Apoptosis.
Cell(Cambridge,Mass.), 152, 2013
|
|
4BD8
 
 | | Bax domain swapped dimer induced by BimBH3 with CHAPS | | Descriptor: | 1,2-ETHANEDIOL, APOPTOSIS REGULATOR BAX, PRASEODYMIUM ION | | Authors: | Czabotar, P.E, Westphal, D, Adams, J.M, Colman, P.M. | | Deposit date: | 2012-10-05 | | Release date: | 2013-02-13 | | Last modified: | 2024-05-01 | | Method: | X-RAY DIFFRACTION (2.22 Å) | | Cite: | Bax Crystal Structures Reveal How Bh3 Domains Activate Bax and Nucleate its Oligomerization to Induce Apoptosis.
Cell(Cambridge,Mass.), 152, 2013
|
|
4BDU
 
 | | Bax BH3-in-Groove dimer (GFP) | | Descriptor: | GREEN FLUORESCENT PROTEIN, APOPTOSIS REGULATOR BAX | | Authors: | Czabotar, P.E, Colman, P.M. | | Deposit date: | 2012-10-08 | | Release date: | 2013-02-13 | | Last modified: | 2019-10-23 | | Method: | X-RAY DIFFRACTION (2.998 Å) | | Cite: | Bax Crystal Structures Reveal How Bh3 Domains Activate Bax and Nucleate its Oligomerization to Induce Apoptosis.
Cell(Cambridge,Mass.), 152, 2013
|
|
7KIA
 
 | | Crystal structure of FGFR2 kinase domain gatekeeper mutant V564F in complex with covalent compound 19 | | Descriptor: | 1-[4-(4-{4-(4-methylpiperazin-1-yl)-6-[(3-methyl-1H-pyrazol-5-yl)amino]pyrimidin-2-yl}phenyl)piperidin-1-yl]prop-2-en-1-one, CITRATE ANION, Fibroblast growth factor receptor 2, ... | | Authors: | Ke, J, Wibowo, A.S, Carter, J.J, Larsen, N.A. | | Deposit date: | 2020-10-23 | | Release date: | 2021-02-10 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (2.22 Å) | | Cite: | Discovery of Aminopyrazole Derivatives as Potent Inhibitors of Wild-Type and Gatekeeper Mutant FGFR2 and 3.
Acs Med.Chem.Lett., 12, 2021
|
|
7KIE
 
 | | Crystal structure of FGFR2 kinase domain gatekeeper mutant V564F in complex with covalent compound 3 | | Descriptor: | CITRATE ANION, Fibroblast growth factor receptor 2, GLYCEROL, ... | | Authors: | Ke, J, Wibowo, A.S, Carter, J.J, Larsen, N.A. | | Deposit date: | 2020-10-23 | | Release date: | 2021-02-10 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (2.47 Å) | | Cite: | Discovery of Aminopyrazole Derivatives as Potent Inhibitors of Wild-Type and Gatekeeper Mutant FGFR2 and 3.
Acs Med.Chem.Lett., 12, 2021
|
|
