1XIP
 
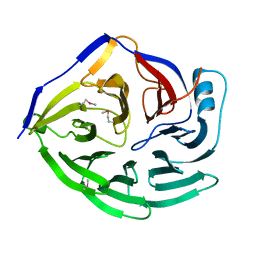 | |
7PER
 
 | | Model of the inner ring of the human nuclear pore complex | | Descriptor: | Nuclear pore complex protein Nup155, Nuclear pore complex protein Nup205, Nuclear pore complex protein Nup93, ... | | Authors: | Schuller, A.P, Wojtynek, M, Mankus, D, Tatli, M, Kronenberg-Tenga, R, Regmi, S.G, Dasso, M, Weis, K, Medalia, O, Schwartz, T.U. | | Deposit date: | 2021-08-11 | | Release date: | 2021-10-20 | | Last modified: | 2024-10-16 | | Method: | ELECTRON MICROSCOPY (35 Å) | | Cite: | The cellular environment shapes the nuclear pore complex architecture.
Nature, 598, 2021
|
|
7PEQ
 
 | | Model of the outer rings of the human nuclear pore complex | | Descriptor: | Nuclear pore complex protein Nup107, Nuclear pore complex protein Nup133, Nuclear pore complex protein Nup160, ... | | Authors: | Schuller, A.P, Wojtynek, M, Mankus, D, Tatli, M, Kronenberg-Tenga, R, Regmi, S.G, Dasso, M, Weis, K, Medalia, O, Schwartz, T.U. | | Deposit date: | 2021-08-11 | | Release date: | 2021-10-20 | | Last modified: | 2024-07-17 | | Method: | ELECTRON MICROSCOPY (35 Å) | | Cite: | The cellular environment shapes the nuclear pore complex architecture.
Nature, 598, 2021
|
|
1QGR
 
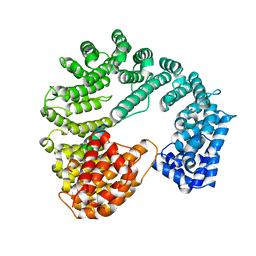 | | STRUCTURE OF IMPORTIN BETA BOUND TO THE IBB DOMAIN OF IMPORTIN ALPHA (II CRYSTAL FORM, GROWN AT LOW PH) | | Descriptor: | PROTEIN (IMPORTIN ALPHA-2 SUBUNIT), PROTEIN (IMPORTIN BETA SUBUNIT) | | Authors: | Cingolani, G, Petosa, C, Weis, K, Muller, C.W. | | Deposit date: | 1999-05-04 | | Release date: | 1999-05-24 | | Last modified: | 2023-08-16 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Structure of importin-beta bound to the IBB domain of importin-alpha.
Nature, 399, 1999
|
|
1QGK
 
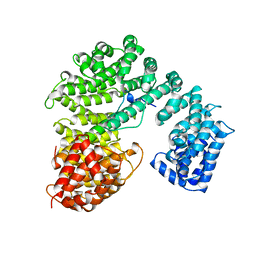 | | STRUCTURE OF IMPORTIN BETA BOUND TO THE IBB DOMAIN OF IMPORTIN ALPHA | | Descriptor: | PROTEIN (IMPORTIN ALPHA-2 SUBUNIT), PROTEIN (IMPORTIN BETA SUBUNIT) | | Authors: | Cingolani, G, Petosa, C, Weis, K, Muller, C.W. | | Deposit date: | 1999-04-29 | | Release date: | 1999-05-24 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Structure of importin-beta bound to the IBB domain of importin-alpha.
Nature, 399, 1999
|
|
6ZL2
 
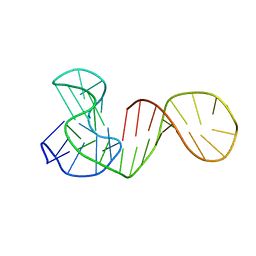 | |
6ERL
 
 | |
6EQY
 
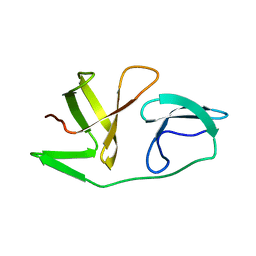 | | 4th KOW domain of human hSpt5 | | Descriptor: | Transcription elongation factor SPT5 | | Authors: | Zuber, P.K, Schweimer, K, Roesch, P, Woehrl, B.M. | | Deposit date: | 2017-10-16 | | Release date: | 2018-10-17 | | Last modified: | 2024-05-15 | | Method: | SOLUTION NMR | | Cite: | Structure and nucleic acid binding properties of KOW domains 4 and 6-7 of human transcription elongation factor DSIF.
Sci Rep, 8, 2018
|
|
1E09
 
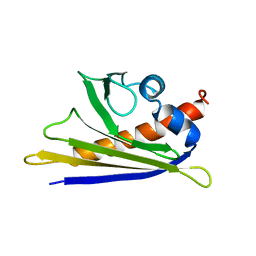 | | Solution Structure of the Major Cherry Allergen Pru av 1 | | Descriptor: | PRU AV 1 | | Authors: | Neudecker, P, Nerkamp, J, Schweimer, K, Sticht, H, Boehm, M, Scheurer, S, Vieths, S, Roesch, P. | | Deposit date: | 2000-03-15 | | Release date: | 2001-03-15 | | Last modified: | 2024-05-15 | | Method: | SOLUTION NMR | | Cite: | Allergic Cross-Reactivity Made Visible: The Solution Structure of the Major Cherry Allergen Pru Av 1
J.Biol.Chem., 276, 2001
|
|
3GFP
 
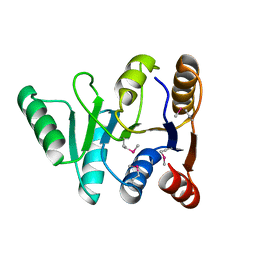 | | Structure of the C-terminal domain of the DEAD-box protein Dbp5 | | Descriptor: | DEAD box protein 5 | | Authors: | Erzberger, J.P, Dossani, Z.Y, Weirich, C.S, Weis, K, Berger, J.M. | | Deposit date: | 2009-02-27 | | Release date: | 2009-09-01 | | Last modified: | 2024-10-09 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Structure of the C-terminus of the mRNA export factor Dbp5 reveals the interaction surface for the ATPase activator Gle1
Proc.Natl.Acad.Sci.USA, 106, 2009
|
|
8QKX
 
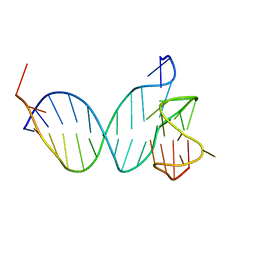 | |
1E10
 
 | | [2Fe-2S]-Ferredoxin from Halobacterium salinarum | | Descriptor: | ACETYL GROUP, FE2/S2 (INORGANIC) CLUSTER, FERREDOXIN | | Authors: | Marg, B.-L, Schweimer, K, Oesterhelt, D, Roesch, P, Sticht, H. | | Deposit date: | 2000-04-12 | | Release date: | 2001-04-09 | | Last modified: | 2011-07-13 | | Method: | SOLUTION NMR | | Cite: | A Two-Alpha-Helix Extra Domain Mediates the Halophilic Character of a Plant-Type Ferredoxin from Halophilic Archaea.
Biochemistry, 44, 2005
|
|
8F60
 
 | | anti-BTLA monoclonal antibody r23C8 in complex with BTLA | | Descriptor: | ACETATE ION, B- and T-lymphocyte attenuator, alpha-D-mannopyranose-(1-6)-beta-D-mannopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, ... | | Authors: | Hendle, J, Atwell, S, Lieu, R, Hickey, M, Weichert, K. | | Deposit date: | 2022-11-15 | | Release date: | 2023-05-24 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (1.64 Å) | | Cite: | Epitope topography of agonist antibodies to the checkpoint inhibitory receptor BTLA.
Structure, 31, 2023
|
|
8F6O
 
 | | anti-BTLA monoclonal antibody h22B3 in complex with BTLA | | Descriptor: | B- and T-lymphocyte attenuator, h22B3 Fab heavy chain, h22B3 Fab light chain | | Authors: | Hendle, J, Atwell, S, Lieu, R, Hickey, M, Weichert, K. | | Deposit date: | 2022-11-16 | | Release date: | 2023-05-24 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (2.31 Å) | | Cite: | Epitope topography of agonist antibodies to the checkpoint inhibitory receptor BTLA.
Structure, 31, 2023
|
|
8F6L
 
 | | anti-BTLA monoclonal antibody h25F7 in complex with BTLA | | Descriptor: | B- and T-lymphocyte attenuator, h25F7 Fab heavy chain, h25F7 Fab light chain | | Authors: | Hendle, J, Atwell, S, Lieu, R, Hickey, M, Weichert, K. | | Deposit date: | 2022-11-16 | | Release date: | 2023-05-24 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (1.85 Å) | | Cite: | Epitope topography of agonist antibodies to the checkpoint inhibitory receptor BTLA.
Structure, 31, 2023
|
|
1H2O
 
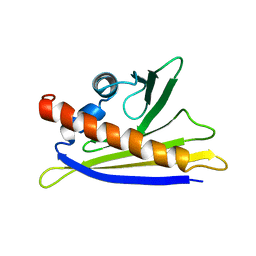 | | SOLUTION STRUCTURE OF THE MAJOR CHERRY ALLERGEN PRU AV 1 MUTANT E45W | | Descriptor: | MAJOR ALLERGEN PRU AV 1 | | Authors: | Neudecker, P, Lehmann, K, Nerkamp, J, Schweimer, K, Sticht, H, Boehm, M, Scheurer, S, Vieths, S, Roesch, P. | | Deposit date: | 2002-08-12 | | Release date: | 2003-08-28 | | Last modified: | 2024-05-15 | | Method: | SOLUTION NMR | | Cite: | Mutational Epitope Analysis of Pru Av 1 and Api G 1, the Major Allergens of Cherry (Prunus Avium) and Celery (Apium Graveolens): Correlating Ige Reactivity with Three-Dimensional Structure
Biochem.J., 376, 2003
|
|
6R9L
 
 | |
6R9K
 
 | |
6U07
 
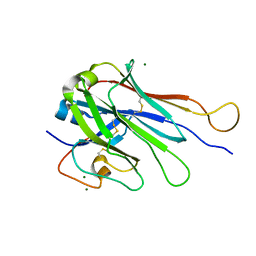 | | Computational Stabilization of T Cell Receptor Constant Domains | | Descriptor: | MAGNESIUM ION, Stabilized T cell receptor constant domain (Calpha), Stabilized T cell receptor constant domain (Cbeta) | | Authors: | Froning, K, Maguire, J, Sereno, A, Huang, F, Chang, S, Weichert, K, Frommelt, A.J, Dong, J, Wu, X, Austin, H, Conner, E.M, Fitchett, J.R, Heng, A.R, Balasubramaniam, D, Hilgers, M.T, Kuhlman, B, Demarest, S.J. | | Deposit date: | 2019-08-13 | | Release date: | 2020-04-15 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (1.76 Å) | | Cite: | Computational stabilization of T cell receptors allows pairing with antibodies to form bispecifics.
Nat Commun, 11, 2020
|
|
1E4T
 
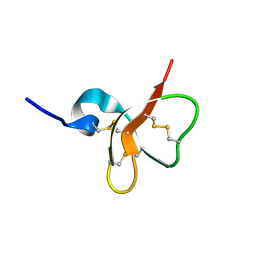 | | Solution structure of the mouse defensin mBD-7 | | Descriptor: | Beta-defensin 7 | | Authors: | Bauer, F, Schweimer, K, Kluver, E, Adermann, K, Forssmann, W.G, Roesch, P, Sticht, H. | | Deposit date: | 2000-07-12 | | Release date: | 2001-07-12 | | Last modified: | 2024-10-16 | | Method: | SOLUTION NMR | | Cite: | Structure determination of human and murine beta-defensins reveals structural conservation in the absence of significant sequence similarity.
Protein Sci., 10, 2001
|
|
1E4Q
 
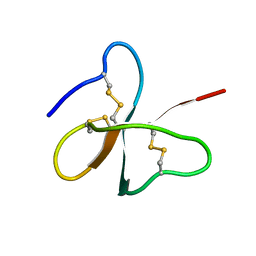 | | Solution structure of the human defensin hBD-2 | | Descriptor: | BETA-DEFENSIN 2 | | Authors: | Bauer, F, Schweimer, K, Kluver, E, Adermann, K, Forssmann, W.G, Roesch, P, Sticht, H. | | Deposit date: | 2000-07-12 | | Release date: | 2001-07-12 | | Last modified: | 2024-10-16 | | Method: | SOLUTION NMR | | Cite: | Structure Determination of Human and Murine Beta-Defensins Reveals Structural Conservation in the Absence of Significant Sequence Similarity
Protein Sci., 10, 2001
|
|
1E4R
 
 | | Solution structure of the mouse defensin mBD-8 | | Descriptor: | Beta-defensin 8 | | Authors: | Bauer, F, Schweimer, K, Kluver, E, Adermann, K, Forssmann, W.G, Roesch, P, Sticht, H. | | Deposit date: | 2000-07-12 | | Release date: | 2001-07-12 | | Last modified: | 2018-06-20 | | Method: | SOLUTION NMR | | Cite: | Structure determination of human and murine beta-defensins reveals structural conservation in the absence of significant sequence similarity.
Protein Sci., 10, 2001
|
|
1E4S
 
 | | Solution structure of the human defensin hBD-1 | | Descriptor: | BETA-DEFENSIN 1 | | Authors: | Bauer, F, Schweimer, K, Kluver, E, Adermann, K, Forssmann, W.G, Roesch, P, Sticht, H. | | Deposit date: | 2000-07-12 | | Release date: | 2001-07-12 | | Last modified: | 2011-07-13 | | Method: | SOLUTION NMR | | Cite: | Structure Determination of Human and Murine Beta-Defensins Reveals Structural Conservation in the Absence of Significant Sequence Similarity
Protein Sci., 10, 2001
|
|
3PEU
 
 | | S. cerevisiae Dbp5 L327V C-terminal domain bound to Gle1 H337R and IP6 | | Descriptor: | ATP-dependent RNA helicase DBP5, GLYCEROL, INOSITOL HEXAKISPHOSPHATE, ... | | Authors: | Montpetit, B, Thomsen, N.D, Helmke, K.J, Seeliger, M.A, Berger, J.M, Weis, K. | | Deposit date: | 2010-10-27 | | Release date: | 2011-03-23 | | Last modified: | 2020-10-14 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | A conserved mechanism of DEAD-box ATPase activation by nucleoporins and InsP(6) in mRNA export.
Nature, 472, 2011
|
|
3PEW
 
 | | S. cerevisiae Dbp5 L327V bound to RNA and ADP BeF3 | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, ATP-dependent RNA helicase DBP5, BERYLLIUM TRIFLUORIDE ION, ... | | Authors: | Montpetit, B, Thomsen, N.D, Helmke, K.J, Seeliger, M.A, Berger, J.M, Weis, K. | | Deposit date: | 2010-10-27 | | Release date: | 2011-03-23 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (1.501 Å) | | Cite: | A conserved mechanism of DEAD-box ATPase activation by nucleoporins and InsP(6) in mRNA export.
Nature, 472, 2011
|
|
