7XE1
 
 | | Crystal structure of LSD2 in complex with cis-4-Br-PCPA | | Descriptor: | 3-(4-bromophenyl)propanal, CITRATE ANION, DI(HYDROXYETHYL)ETHER, ... | | Authors: | Niwa, H, Sato, S, Umehara, T. | | Deposit date: | 2022-03-29 | | Release date: | 2022-09-14 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2.07 Å) | | Cite: | Structure-Activity Relationship and In Silico Evaluation of cis- and trans-PCPA-Derived Inhibitors of LSD1 and LSD2
Acs Med.Chem.Lett., 13, 2022
|
|
7XE2
 
 | | Crystal structure of LSD2 in complex with trans-4-Br-PCPA | | Descriptor: | 3-(4-bromophenyl)propanal, CITRATE ANION, FLAVIN-ADENINE DINUCLEOTIDE, ... | | Authors: | Niwa, H, Sato, S, Umehara, T. | | Deposit date: | 2022-03-29 | | Release date: | 2022-09-14 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2.05 Å) | | Cite: | Structure-Activity Relationship and In Silico Evaluation of cis- and trans-PCPA-Derived Inhibitors of LSD1 and LSD2
Acs Med.Chem.Lett., 13, 2022
|
|
7XE3
 
 | | Crystal structure of LSD2 in complex with cis-4-Br-2,5-F2-PCPA (S1024) | | Descriptor: | 1,2-ETHANEDIOL, 3-[4-bromanyl-2,5-bis(fluoranyl)phenyl]propanal, CITRATE ANION, ... | | Authors: | Niwa, H, Sato, S, Umehara, T. | | Deposit date: | 2022-03-29 | | Release date: | 2022-09-14 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2.82 Å) | | Cite: | Structure-Activity Relationship and In Silico Evaluation of cis- and trans-PCPA-Derived Inhibitors of LSD1 and LSD2
Acs Med.Chem.Lett., 13, 2022
|
|
5YVB
 
 | | Structure of CaMKK2 in complex with CKI-011 | | Descriptor: | (3Z)-5-chloro-3-[(1-methyl-1H-pyrazol-4-yl)methylidene]-1,3-dihydro-2H-indol-2-one, CHLORIDE ION, Calcium/calmodulin-dependent protein kinase kinase 2, ... | | Authors: | Niwa, H, Handa, N, Yokoyama, S. | | Deposit date: | 2017-11-24 | | Release date: | 2018-12-05 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (2.02 Å) | | Cite: | Protein ligand interaction analysis against new CaMKK2 inhibitors by use of X-ray crystallography and the fragment molecular orbital (FMO) method.
J.Mol.Graph.Model., 99, 2020
|
|
5YV9
 
 | | Structure of CaMKK2 in complex with CKI-009 | | Descriptor: | 5-chloro-2-methoxy-4[(1Z)-3-(4-methoxyphenyl)-3-oxoprop-1-en-1-yl]aminobenzoic acid, CHLORIDE ION, Calcium/calmodulin-dependent protein kinase kinase 2, ... | | Authors: | Niwa, H, Handa, N, Yokoyama, S. | | Deposit date: | 2017-11-24 | | Release date: | 2018-12-05 | | Last modified: | 2024-10-09 | | Method: | X-RAY DIFFRACTION (2.53 Å) | | Cite: | Protein ligand interaction analysis against new CaMKK2 inhibitors by use of X-ray crystallography and the fragment molecular orbital (FMO) method.
J.Mol.Graph.Model., 99, 2020
|
|
5YV8
 
 | | Structure of CaMKK2 in complex with CKI-002 | | Descriptor: | 1-amino-4-hydroxy-9,10-dioxo-9,10-dihydroanthracene-2-carboxylic acid, CHLORIDE ION, Calcium/calmodulin-dependent protein kinase kinase 2, ... | | Authors: | Niwa, H, Handa, N, Yokoyama, S. | | Deposit date: | 2017-11-24 | | Release date: | 2018-12-05 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (1.927 Å) | | Cite: | Protein ligand interaction analysis against new CaMKK2 inhibitors by use of X-ray crystallography and the fragment molecular orbital (FMO) method.
J.Mol.Graph.Model., 99, 2020
|
|
5YVC
 
 | | Structure of CaMKK2 in complex with CKI-012 | | Descriptor: | 3-{2,4-dimethyl-5-[(Z)-(2-oxo-1,2-dihydro-3H-indol-3-ylidene)methyl]-1H-pyrrol-3-yl}propanoic acid, CHLORIDE ION, Calcium/calmodulin-dependent protein kinase kinase 2, ... | | Authors: | Niwa, H, Handa, N, Yokoyama, S. | | Deposit date: | 2017-11-24 | | Release date: | 2018-12-05 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (2.02 Å) | | Cite: | Protein ligand interaction analysis against new CaMKK2 inhibitors by use of X-ray crystallography and the fragment molecular orbital (FMO) method.
J.Mol.Graph.Model., 99, 2020
|
|
5YVA
 
 | | Structure of CaMKK2 in complex with CKI-010 | | Descriptor: | 3-(1H-tetrazol-5-yl)-10lambda~6~-thioxanthene-9,10,10-trione, CHLORIDE ION, Calcium/calmodulin-dependent protein kinase kinase 2, ... | | Authors: | Niwa, H, Handa, N, Yokoyama, S. | | Deposit date: | 2017-11-24 | | Release date: | 2018-12-05 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (2.574 Å) | | Cite: | Protein ligand interaction analysis against new CaMKK2 inhibitors by use of X-ray crystallography and the fragment molecular orbital (FMO) method.
J.Mol.Graph.Model., 99, 2020
|
|
1V7A
 
 | | Crystal structures of adenosine deaminase complexed with potent inhibitors | | Descriptor: | 1-{(1R,2S)-2-HYDROXY-1-[2-(2-NAPHTHYLOXY)ETHYL]PROPYL}-1H-IMIDAZONE-4-CARBOXAMIDE, ZINC ION, adenosine deaminase | | Authors: | Kinoshita, T. | | Deposit date: | 2003-12-14 | | Release date: | 2004-12-21 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Structure-based design and synthesis of non-nucleoside, potent, and orally bioavailable adenosine deaminase inhibitors
J.Med.Chem., 47, 2004
|
|
1V79
 
 | | Crystal structures of adenosine deaminase complexed with potent inhibitors | | Descriptor: | 1-{(1R,2S)-1-[2-(2,3,-DICHLOROPHENYL)ETHYL]-2-HYDROXYPROPYL}-1H-IMIDAZOLE-4-CARBOXAMIDE, Adenosine deaminase, ZINC ION | | Authors: | Kinoshita, T. | | Deposit date: | 2003-12-14 | | Release date: | 2004-12-21 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Structure-based design and synthesis of non-nucleoside, potent, and orally bioavailable adenosine deaminase inhibitors
J.Med.Chem., 47, 2004
|
|
4ZTD
 
 | | Crystal Structure of Human PCNA in complex with a TRAIP peptide | | Descriptor: | ALA-GLY-ALA-GLY-ALA, ALA-PHE-GLN-ALA-LYS-LEU-ASP-THR-PHE-LEU-TRP-SER, Proliferating cell nuclear antigen | | Authors: | Montoya, G, Mortuza, G.B, Blanco, F.J, Ibanez de Opakua, A. | | Deposit date: | 2015-05-14 | | Release date: | 2015-12-16 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (2.199 Å) | | Cite: | TRAIP is a PCNA-binding ubiquitin ligase that protects genome stability after replication stress.
J.Cell Biol., 212, 2016
|
|
6IO0
 
 | | Human IDH1 R132C mutant complexed with compound A. | | Descriptor: | (2E)-3-{3-[3-(2,6-dichlorophenyl)-5-(propan-2-yl)-1,2-oxazole-4-carbonyl]-1-methyl-1H-indol-7-yl}prop-2-enoic acid, CITRIC ACID, GLYCEROL, ... | | Authors: | Suzuki, M, Baba, D, Hanzawa, H. | | Deposit date: | 2018-10-29 | | Release date: | 2019-10-30 | | Last modified: | 2024-03-27 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | A Potent Blood-Brain Barrier-Permeable Mutant IDH1 Inhibitor Suppresses the Growth of Glioblastoma with IDH1 Mutation in a Patient-Derived Orthotopic Xenograft Model.
Mol.Cancer Ther., 19, 2020
|
|
7CD7
 
 | | GFP-40/GFPuv complex, Form I | | Descriptor: | GFP-40, Green fluorescent protein | | Authors: | Yasui, N, Yamashita, A. | | Deposit date: | 2020-06-18 | | Release date: | 2021-01-13 | | Last modified: | 2024-10-09 | | Method: | X-RAY DIFFRACTION (1.704 Å) | | Cite: | A sweet protein monellin as a non-antibody scaffold for synthetic binding proteins.
J.Biochem., 169, 2021
|
|
7CD8
 
 | | GFP-40/GFPuv complex, Form II | | Descriptor: | GFP-40, Green fluorescent protein | | Authors: | Yasui, N, Yamashita, A. | | Deposit date: | 2020-06-18 | | Release date: | 2021-01-13 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | A sweet protein monellin as a non-antibody scaffold for synthetic binding proteins.
J.Biochem., 169, 2021
|
|
2E1W
 
 | | Crystal structure of adenosine deaminase complexed with potent inhibitors | | Descriptor: | 1-{(1R,2S)-2-HYDROXY-1-[2-(1-NAPHTHYL)ETHYL]PROPYL}-1H-IMIDAZOLE-4-CARBOXAMIDE, Adenosine deaminase, ZINC ION | | Authors: | Kinoshita, T. | | Deposit date: | 2006-10-30 | | Release date: | 2006-11-07 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Structure-Based Design and Synthesis of Non-Nucleoside, Potent, and Orally Bioavailable Adenosine Deaminase Inhibitors
J.Med.Chem., 47, 2004
|
|
7VQT
 
 | | Crystal structure of LSD1 in complex with compound 5 | | Descriptor: | 3-[3,5-bis(fluoranyl)-2-[(2-fluoranylpyridin-4-yl)methoxy]phenyl]propanal, FLAVIN-ADENINE DINUCLEOTIDE, GLYCEROL, ... | | Authors: | Niwa, H, Koda, Y, Sato, S, Yamamoto, H, Koyama, H, Umehara, T. | | Deposit date: | 2021-10-20 | | Release date: | 2022-06-01 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2.91 Å) | | Cite: | Design and Synthesis of Tranylcypromine-Derived LSD1 Inhibitors with Improved hERG and Microsomal Stability Profiles.
Acs Med.Chem.Lett., 13, 2022
|
|
7VQS
 
 | | Crystal structure of LSD1 in complex with compound 4 | | Descriptor: | 3-[3,5-bis(fluoranyl)-2-[(2-fluoranylpyridin-3-yl)methoxy]phenyl]propanal, FLAVIN-ADENINE DINUCLEOTIDE, GLYCEROL, ... | | Authors: | Niwa, H, Koda, Y, Sato, S, Yamamoto, H, Koyama, H, Umehara, T. | | Deposit date: | 2021-10-20 | | Release date: | 2022-06-01 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2.94 Å) | | Cite: | Design and Synthesis of Tranylcypromine-Derived LSD1 Inhibitors with Improved hERG and Microsomal Stability Profiles.
Acs Med.Chem.Lett., 13, 2022
|
|
7VQU
 
 | | Crystal structure of LSD1 in complex with compound S1427 | | Descriptor: | 3-[3,5-bis(fluoranyl)-2-[(2-fluoranylpyridin-3-yl)methoxy]phenyl]propanal, FLAVIN-ADENINE DINUCLEOTIDE, GLYCEROL, ... | | Authors: | Niwa, H, Koda, Y, Sato, S, Yamamoto, H, Koyama, H, Umehara, T. | | Deposit date: | 2021-10-20 | | Release date: | 2022-06-01 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2.94 Å) | | Cite: | Design and Synthesis of Tranylcypromine-Derived LSD1 Inhibitors with Improved hERG and Microsomal Stability Profiles.
Acs Med.Chem.Lett., 13, 2022
|
|
2DS1
 
 | | Human cyclin dependent kinase 2 complexed with the CDK4 inhibitor | | Descriptor: | (13R,15S)-13-METHYL-16-OXA-8,9,12,22,24-PENTAAZAHEXACYCLO[15.6.2.16,9.1,12,15.0,2,7.0,21,25]HEPTACOSA-1(24),2,4,6,17(25 ),18,20-HEPTAENE-23,26-DIONE, Cell division protein kinase 2 | | Authors: | Ikuta, M. | | Deposit date: | 2006-06-17 | | Release date: | 2007-06-19 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Structure-based drug design of a highly potent CDK1,2,4,6 inhibitor with novel macrocyclic quinoxalin-2-one structure
Bioorg.Med.Chem.Lett., 16, 2006
|
|
1LRA
 
 | | CRYSTALLOGRAPHIC STUDY OF GLU 58 ALA RNASE T1(ASTERISK)2'-GUANOSINE MONOPHOSPHATE AT 1.9 ANGSTROMS RESOLUTION | | Descriptor: | GUANOSINE-2'-MONOPHOSPHATE, RIBONUCLEASE T1, SODIUM ION | | Authors: | Pletinckx, J, Steyaert, J, Choe, H.-W, Heinemann, U, Wyns, L. | | Deposit date: | 1993-10-01 | | Release date: | 1994-01-31 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Crystallographic study of Glu58Ala RNase T1 x 2'-guanosine monophosphate at 1.9-A resolution.
Biochemistry, 33, 1994
|
|
1GOY
 
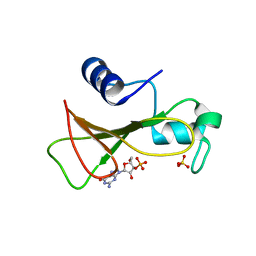 | | HYDROLASE(ENDORIBONUCLEASE)RIBONUCLEASE BI(G SPECIFIC ENDONUCLEASE) (E.C.3.1.27.-) COMPLEXED WITH GUANOSINE-3'-PHOSPHATE (3'-GMP) | | Descriptor: | GUANOSINE-3'-MONOPHOSPHATE, RIBONUCLEASE, SULFATE ION | | Authors: | Polyakov, K.M, Lebedev, A.A, Pavlovsky, A.G, Sanishvili, R.G, Dodson, G.G. | | Deposit date: | 2001-10-26 | | Release date: | 2001-11-29 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | The Structure of Substrate-Free Microbial Ribonuclease Binase and of its Complexes with 3'Gmp and Sulfate Ions
Acta Crystallogr.,Sect.D, 58, 2002
|
|
1RNT
 
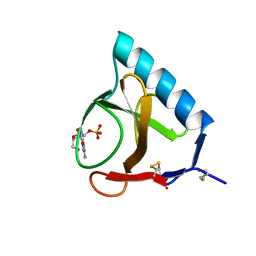 | | RESTRAINED LEAST-SQUARES REFINEMENT OF THE CRYSTAL STRUCTURE OF THE RIBONUCLEASE T1(ASTERISK)2(PRIME)-GUANYLIC ACID COMPLEX AT 1.9 ANGSTROMS RESOLUTION | | Descriptor: | GUANOSINE-2'-MONOPHOSPHATE, RIBONUCLEASE T1 ISOZYME | | Authors: | Saenger, W, Arni, R, Heinemann, U, Tokuoka, R. | | Deposit date: | 1987-07-10 | | Release date: | 1987-10-16 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Restrained Least-Squares Refinement of the Crystal Structure of the Ribonuclease T1(Asterisk)2(Prime)-Guanylic Acid Complex at 1.9 Angstroms Resolution
Acta Crystallogr.,Sect.B, 43, 1987
|
|
2Z4R
 
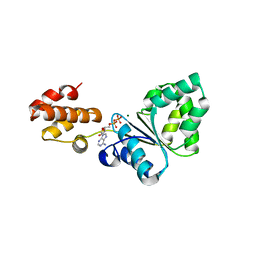 | | Crystal structure of domain III from the Thermotoga maritima replication initiation protein DnaA | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, Chromosomal replication initiator protein dnaA, MAGNESIUM ION | | Authors: | Fujikawa, N, Ozaki, S, Kagawa, W, Park, S.-Y, Katayama, T, Kurumizaka, H, Yokoyama, S, RIKEN Structural Genomics/Proteomics Initiative (RSGI) | | Deposit date: | 2007-06-25 | | Release date: | 2008-02-19 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (3.05 Å) | | Cite: | A Common Mechanism for the ATP-DnaA-dependent Formation of Open Complexes at the Replication Origin
J.Biol.Chem., 283, 2008
|
|
2Z4S
 
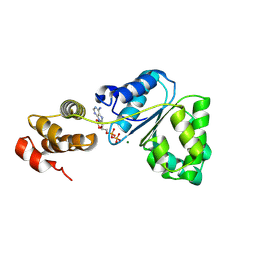 | | Crystal structure of domain III from the Thermotoga maritima replication initiation protein DnaA | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, Chromosomal replication initiator protein dnaA, MAGNESIUM ION | | Authors: | Fujikawa, N, Ozaki, S, Kagawa, W, Park, S.-Y, Katayama, T, Kurumizaka, H, Yokoyama, S, RIKEN Structural Genomics/Proteomics Initiative (RSGI) | | Deposit date: | 2007-06-25 | | Release date: | 2008-02-19 | | Last modified: | 2024-08-14 | | Method: | X-RAY DIFFRACTION (3 Å) | | Cite: | A Common Mechanism for the ATP-DnaA-dependent Formation of Open Complexes at the Replication Origin.
J.Biol.Chem., 283, 2008
|
|
