6WG7
 
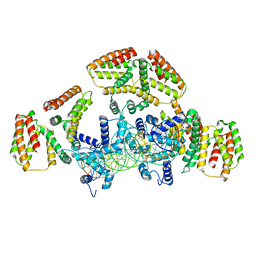 | | Coordinates of NanR dimer fitted in Hexameric NanR-DNA hetero-complex cryo-EM map | | Descriptor: | DNA (35-MER), HTH-type transcriptional repressor NanR | | Authors: | Hariprasad, V, Horne, C, Santosh, P, Amy, H, Emre, B, Rachel, N, Michael, G, Georg, R, Borries, D, Renwick, D. | | Deposit date: | 2020-04-05 | | Release date: | 2021-03-10 | | Last modified: | 2024-03-06 | | Method: | ELECTRON MICROSCOPY (8.3 Å) | | Cite: | Mechanism of NanR gene repression and allosteric induction of bacterial sialic acid metabolism.
Nat Commun, 12, 2021
|
|
6WFQ
 
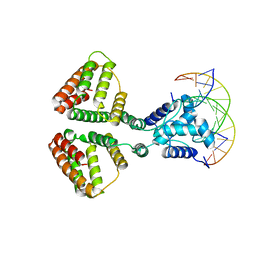 | | NanR dimer-DNA hetero-complex | | Descriptor: | DNA (5'-D(P*GP*GP*TP*AP*TP*AP*AP*CP*AP*GP*GP*TP*AP*TP*A)-3'), DNA (5'-D(P*TP*AP*TP*AP*CP*CP*TP*GP*TP*TP*AP*TP*AP*CP*C)-3'), HTH-type transcriptional repressor NanR | | Authors: | Hariprasad, V, Horne, C, Santosh, P, Amy, H, Emre, B, Rachel, N, Michael, G, Georg, R, Borries, D, Renwick, D. | | Deposit date: | 2020-04-03 | | Release date: | 2021-03-10 | | Last modified: | 2024-03-06 | | Method: | ELECTRON MICROSCOPY (3.9 Å) | | Cite: | Mechanism of NanR gene repression and allosteric induction of bacterial sialic acid metabolism.
Nat Commun, 12, 2021
|
|
2QST
 
 | |
2FP3
 
 | |
6U6Y
 
 | | Human SAMHD1 bound to ribo(CGCCU)-oligonucleotide | | Descriptor: | 2'-DEOXYADENOSINE 5'-TRIPHOSPHATE, Deoxynucleoside triphosphate triphosphohydrolase SAMHD1, RNA CGCCU, ... | | Authors: | Taylor, A.B, Bhattacharya, A, Wang, Z, Ivanov, D.N. | | Deposit date: | 2019-08-30 | | Release date: | 2020-09-02 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (2.47 Å) | | Cite: | Nucleic acid binding by SAMHD1 contributes to the antiretroviral activity and is enhanced by the GpsN modification.
Nat Commun, 12, 2021
|
|
6DR6
 
 | |
5FR6
 
 | | The structure of polycomb ULD complex | | Descriptor: | POLYCOMB COMPLEX PROTEIN BMI-1 | | Authors: | Gray, F, Cho, H.J, Cierpicki, T. | | Deposit date: | 2015-12-15 | | Release date: | 2016-11-16 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (2.51 Å) | | Cite: | Bmi1 Regulates Prc1 Architecture and Activity Through Homo-and Hetero-Oligomerization
Nat.Commun., 7, 2016
|
|
4WYM
 
 | | Structural basis of HIV-1 capsid recognition by CPSF6 | | Descriptor: | Capsid protein p24, ISOFORM 2 OF CLEAVAGE AND POLYADENYLATION SPECIFICITY FACTOR SUBUNIT 6 | | Authors: | Battacharya, A, Taylor, A.B, Hart, P.J, Ivanov, D.N. | | Deposit date: | 2014-11-17 | | Release date: | 2014-12-17 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Structural basis of HIV-1 capsid recognition by PF74 and CPSF6.
Proc.Natl.Acad.Sci.USA, 111, 2014
|
|
8THI
 
 | | Cryo-EM structure of the Tripartite ATP-independent Periplasmic (TRAP) transporter SiaQM from Haemophilus influenzae (parallel dimer) | | Descriptor: | (1S)-2-{[{[(2R)-2,3-DIHYDROXYPROPYL]OXY}(HYDROXY)PHOSPHORYL]OXY}-1-[(PALMITOYLOXY)METHYL]ETHYL STEARATE, SODIUM ION, Sialic acid TRAP transporter permease protein SiaT | | Authors: | Davies, J.S, Currie, M.C, Dobson, R.C.J, North, R.A. | | Deposit date: | 2023-07-16 | | Release date: | 2023-11-22 | | Last modified: | 2024-02-28 | | Method: | ELECTRON MICROSCOPY (3.36 Å) | | Cite: | Structural and biophysical analysis of a Haemophilus influenzae tripartite ATP-independent periplasmic (TRAP) transporter.
Elife, 12, 2024
|
|
8THJ
 
 | | Cryo-EM structure of the Tripartite ATP-independent Periplasmic (TRAP) transporter SiaQM from Haemophilus influenzae (antiparallel dimer) | | Descriptor: | (1S)-2-{[{[(2R)-2,3-DIHYDROXYPROPYL]OXY}(HYDROXY)PHOSPHORYL]OXY}-1-[(PALMITOYLOXY)METHYL]ETHYL STEARATE, PHOSPHATIDYLETHANOLAMINE, SODIUM ION, ... | | Authors: | Davies, J.S, Currie, M.C, Dobson, R.C.J, North, R.A. | | Deposit date: | 2023-07-16 | | Release date: | 2023-11-22 | | Last modified: | 2024-02-28 | | Method: | ELECTRON MICROSCOPY (2.99 Å) | | Cite: | Structural and biophysical analysis of a Haemophilus influenzae tripartite ATP-independent periplasmic (TRAP) transporter.
Elife, 12, 2024
|
|
5TX4
 
 | | Derivative of mouse TGF-beta2, with a deletion of residues 52-71 and K25R, R26K, L51R, A74K, C77S, L89V, I92V, K94R T95K, I98V single amino acid substitutions, bound to human TGF-beta type II receptor ectodomain residues 15-130 | | Descriptor: | TGF-beta receptor type-2, Transforming growth factor beta-2 | | Authors: | Hinck, A.P, Kim, S. | | Deposit date: | 2016-11-15 | | Release date: | 2017-03-01 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (1.876 Å) | | Cite: | An engineered transforming growth factor beta (TGF-beta ) monomer that functions as a dominant negative to block TGF-beta signaling.
J. Biol. Chem., 292, 2017
|
|
5TX6
 
 | |
5TX2
 
 | | Miniature TGF-beta2 3-mutant monomer | | Descriptor: | Transforming growth factor beta-2 | | Authors: | Taylor, A.B, Kim, S.K, Hart, P.J, Hinck, A.P. | | Deposit date: | 2016-11-15 | | Release date: | 2017-03-01 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (1.82 Å) | | Cite: | An engineered transforming growth factor beta (TGF-beta ) monomer that functions as a dominant negative to block TGF-beta signaling.
J. Biol. Chem., 292, 2017
|
|
6U6Z
 
 | | Human SAMHD1 bound to deoxyribo(TG*TTCA)-oligonucleotide | | Descriptor: | DNA polymer TG(PST)TCA, Deoxynucleoside triphosphate triphosphohydrolase SAMHD1, ZINC ION | | Authors: | Taylor, A.B, Yu, C.H, Ivanov, D.N. | | Deposit date: | 2019-08-30 | | Release date: | 2020-09-02 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Nucleic acid binding by SAMHD1 contributes to the antiretroviral activity and is enhanced by the GpsN modification.
Nat Commun, 12, 2021
|
|
6U6X
 
 | | Human SAMHD1 bound to deoxyribo(C*G*C*C*T)-oligonucleotide | | Descriptor: | DNA SC-GS-SC-SC-DT, Deoxynucleoside triphosphate triphosphohydrolase SAMHD1, ZINC ION | | Authors: | Taylor, A.B, Bhattacharya, A, Wang, Z, Ivanov, D.N. | | Deposit date: | 2019-08-30 | | Release date: | 2020-09-02 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (2.58 Å) | | Cite: | Nucleic acid binding by SAMHD1 contributes to the antiretroviral activity and is enhanced by the GpsN modification.
Nat Commun, 12, 2021
|
|
7M98
 
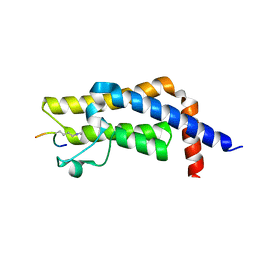 | | ATAD2 bromodomain complexed with histone H4K5ac (res 1-10) ligand | | Descriptor: | ATPase family AAA domain-containing protein 2, Histone H4 | | Authors: | Malone, K.L, Phillips, M, Nix, J.C, Glass, K.C. | | Deposit date: | 2021-03-30 | | Release date: | 2021-09-22 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Coordination of Di-Acetylated Histone Ligands by the ATAD2 Bromodomain.
Int J Mol Sci, 22, 2021
|
|
6I9J
 
 | | Human transforming growth factor beta2 in a tetragonal crystal form | | Descriptor: | Transforming growth factor beta-2 proprotein | | Authors: | Gomis-Ruth, F.X, Marino-Puertas, L, del Amo-Maestro, L, Goulas, T. | | Deposit date: | 2018-11-23 | | Release date: | 2019-07-03 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Recombinant production, purification, crystallization, and structure analysis of human transforming growth factor beta 2 in a new conformation.
Sci Rep, 9, 2019
|
|
3NYJ
 
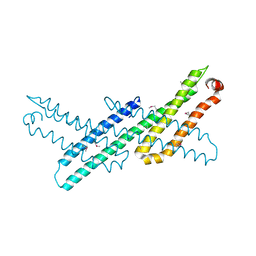 | | Crystal Structure Analysis of APP E2 domain | | Descriptor: | Amyloid beta A4 protein, OSMIUM ION | | Authors: | Ha, Y, Hu, J, Lee, S, Liu, X. | | Deposit date: | 2010-07-15 | | Release date: | 2011-06-01 | | Last modified: | 2017-11-08 | | Method: | X-RAY DIFFRACTION (3.2 Å) | | Cite: | The E2 Domains of APP and APLP1 Share a Conserved Mode of Dimerization.
Biochemistry, 50, 2011
|
|
5JIE
 
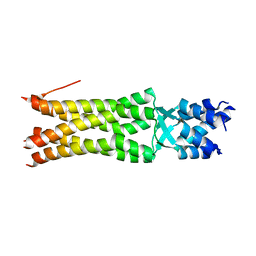 | |
5L1M
 
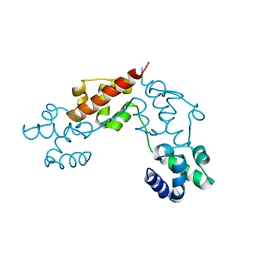 | | CASKIN2 SAM domain tandem | | Descriptor: | Caskin-2 | | Authors: | Donaldson, L.W, Kwan, J.J, Saridakis, V. | | Deposit date: | 2016-07-29 | | Release date: | 2016-08-10 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (2.751 Å) | | Cite: | A new mode of SAM domain mediated oligomerization observed in the CASKIN2 neuronal scaffolding protein.
Cell Commun. Signal, 14, 2016
|
|
3BBD
 
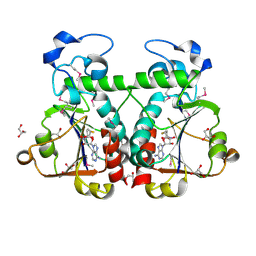 | | M. jannaschii Nep1 complexed with S-adenosyl-homocysteine | | Descriptor: | GLYCEROL, Ribosome biogenesis protein NEP1-like, S-ADENOSYL-L-HOMOCYSTEINE | | Authors: | Taylor, A.B, Meyer, B, Leal, B.Z, Kotter, P, Hart, P.J, Entian, K.-D, Wohnert, J. | | Deposit date: | 2007-11-09 | | Release date: | 2008-02-05 | | Last modified: | 2011-07-13 | | Method: | X-RAY DIFFRACTION (2.15 Å) | | Cite: | The crystal structure of Nep1 reveals an extended SPOUT-class methyltransferase fold and a pre-organized SAM-binding site.
Nucleic Acids Res., 36, 2008
|
|
3BBE
 
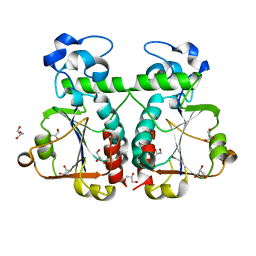 | | M. jannaschii Nep1 | | Descriptor: | GLYCEROL, Ribosome biogenesis protein NEP1-like | | Authors: | Taylor, A.B, Meyer, B, Leal, B.Z, Kotter, P, Hart, P.J, Entian, K.-D, Wohnert, J. | | Deposit date: | 2007-11-09 | | Release date: | 2008-02-05 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | The crystal structure of Nep1 reveals an extended SPOUT-class methyltransferase fold and a pre-organized SAM-binding site.
Nucleic Acids Res., 36, 2008
|
|
3BBH
 
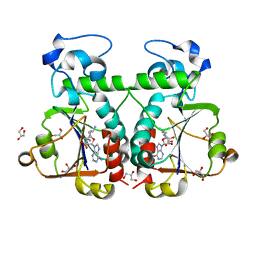 | | M. jannaschii Nep1 complexed with Sinefungin | | Descriptor: | GLYCEROL, Ribosome biogenesis protein NEP1-like, SINEFUNGIN | | Authors: | Taylor, A.B, Meyer, B, Leal, B.Z, Kotter, P, Hart, P.J, Entian, K.-D, Wohnert, J. | | Deposit date: | 2007-11-09 | | Release date: | 2008-02-05 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (2.25 Å) | | Cite: | The crystal structure of Nep1 reveals an extended SPOUT-class methyltransferase fold and a pre-organized SAM-binding site.
Nucleic Acids Res., 36, 2008
|
|
8DFK
 
 | |
8DSS
 
 | |
