8JY8
 
 | |
8P8X
 
 | |
7X05
 
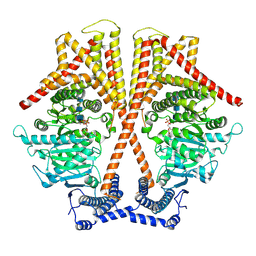 | | CryoEM structure of chitin synthase 1 from Phytophthora sojae complexed with the nascent chitooligosaccharide | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Chitin synthase, MANGANESE (II) ION, ... | | Authors: | Chen, W, Cao, P, Gong, Y, Yang, Q. | | Deposit date: | 2022-02-21 | | Release date: | 2022-09-28 | | Last modified: | 2024-06-26 | | Method: | ELECTRON MICROSCOPY (3.9 Å) | | Cite: | Structural basis for directional chitin biosynthesis.
Nature, 610, 2022
|
|
7X06
 
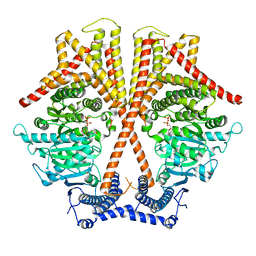 | | CryoEM structure of chitin synthase 1 from Phytophthora sojae complexed with UDP | | Descriptor: | Chitin synthase, MAGNESIUM ION, URIDINE-5'-DIPHOSPHATE | | Authors: | Chen, W, Cao, P, Gong, Y, Yang, Q. | | Deposit date: | 2022-02-21 | | Release date: | 2022-09-28 | | Last modified: | 2024-06-26 | | Method: | ELECTRON MICROSCOPY (3.1 Å) | | Cite: | Structural basis for directional chitin biosynthesis.
Nature, 610, 2022
|
|
7WJM
 
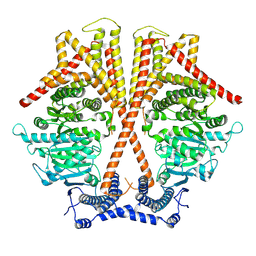 | |
7WJN
 
 | | CryoEM structure of chitin synthase 1 mutant E495A from Phytophthora sojae complexed with UDP-GlcNAc | | Descriptor: | Chitin synthase, MANGANESE (II) ION, URIDINE-DIPHOSPHATE-N-ACETYLGLUCOSAMINE | | Authors: | Chen, W, Cao, P, Gong, Y, Yang, Q. | | Deposit date: | 2022-01-07 | | Release date: | 2022-09-28 | | Last modified: | 2024-06-26 | | Method: | ELECTRON MICROSCOPY (3.3 Å) | | Cite: | Structural basis for directional chitin biosynthesis.
Nature, 610, 2022
|
|
7WJO
 
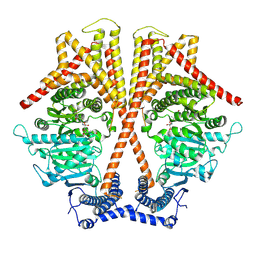 | | CryoEM structure of chitin synthase 1 from Phytophthora sojae complexed with nikkomycin Z | | Descriptor: | (2S)-{[(2S,3S,4S)-2-amino-4-hydroxy-4-(5-hydroxypyridin-2-yl)-3-methylbutanoyl]amino}[(2R,3S,4R,5R)-5-(2,4-dioxo-3,4-dihydropyrimidin-1(2H)-yl)-3,4-dihydroxyoxolan-2-yl]acetic acid (non-preferred name), Chitin synthase | | Authors: | Chen, W, Cao, P, Gong, Y, Yang, Q. | | Deposit date: | 2022-01-07 | | Release date: | 2022-09-28 | | Last modified: | 2024-06-26 | | Method: | ELECTRON MICROSCOPY (3.2 Å) | | Cite: | Structural basis for directional chitin biosynthesis.
Nature, 610, 2022
|
|
6JAX
 
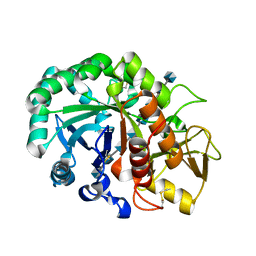 | | Crystal structure of Ostrinia furnacalis Group II chitinase catalytic domain 1 in complex with chitooctaose [(GlcN)8] | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, 2-amino-2-deoxy-beta-D-glucopyranose-(1-4)-2-amino-2-deoxy-beta-D-glucopyranose-(1-4)-2-amino-2-deoxy-beta-D-glucopyranose-(1-4)-2-amino-2-deoxy-beta-D-glucopyranose-(1-4)-2-amino-2-deoxy-beta-D-glucopyranose-(1-4)-2-amino-2-deoxy-beta-D-glucopyranose-(1-4)-2-amino-2-deoxy-beta-D-glucopyranose-(1-4)-2-amino-2-deoxy-beta-D-glucopyranose, Group II chitinase | | Authors: | Chen, W, Zhou, Y, Yang, Q. | | Deposit date: | 2019-01-25 | | Release date: | 2019-05-15 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Structural dissection reveals a general mechanistic principle for group II chitinase (ChtII) inhibition.
J.Biol.Chem., 294, 2019
|
|
6K3Z
 
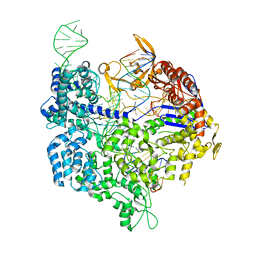 | | Crystal structure of dCas9 in complex with sgRNA and DNA (TGA PAM) | | Descriptor: | CRISPR-associated endonuclease Cas9, DNA (28-MER), DNA (5'-D(*AP*AP*AP*TP*GP*AP*TP*AP*TP*TP*G)-3'), ... | | Authors: | Chen, W, Zhang, H, Zhang, Y, Wang, Y, Gan, J, Ji, Q. | | Deposit date: | 2019-05-22 | | Release date: | 2019-09-25 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (3.2 Å) | | Cite: | Molecular basis for the PAM expansion and fidelity enhancement of an evolved Cas9 nuclease.
Plos Biol., 17, 2019
|
|
6K4P
 
 | | Crystal structure of xCas9 in complex with sgRNA and DNA (TGG PAM) | | Descriptor: | CRISPR-associated endonuclease Cas9/Csn1, DNA (28-MER), PHOSPHATE ION, ... | | Authors: | Chen, W, Zhang, H, Zhang, Y, Wang, Y, Gan, J, Ji, Q. | | Deposit date: | 2019-05-25 | | Release date: | 2019-09-25 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (2.9 Å) | | Cite: | Molecular basis for the PAM expansion and fidelity enhancement of an evolved Cas9 nuclease.
Plos Biol., 17, 2019
|
|
6K4U
 
 | | Crystal structure of xCas9 in complex with sgRNA and DNA (TGA PAM) | | Descriptor: | CRISPR-associated endonuclease Cas9, non-target DNA, sgRNA, ... | | Authors: | Chen, W, Zhang, H, Zhang, Y, Wang, Y, Gan, J, Ji, Q. | | Deposit date: | 2019-05-27 | | Release date: | 2019-09-25 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (3.2 Å) | | Cite: | Molecular basis for the PAM expansion and fidelity enhancement of an evolved Cas9 nuclease.
Plos Biol., 17, 2019
|
|
6K4Q
 
 | | Crystal structure of xCas9 in complex with sgRNA and DNA (CGG PAM) | | Descriptor: | CRISPR-associated endonuclease Cas9/Csn1, DNA (28-MER), DNA (5'-D(*AP*AP*AP*CP*GP*GP*TP*AP*TP*TP*G)-3'), ... | | Authors: | Chen, W, Zhang, H, Zhang, Y, Wang, Y, Gan, J, Ji, Q. | | Deposit date: | 2019-05-26 | | Release date: | 2019-09-25 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | Molecular basis for the PAM expansion and fidelity enhancement of an evolved Cas9 nuclease.
Plos Biol., 17, 2019
|
|
6K4S
 
 | | Crystal structure of xCas9 in complex with sgRNA and DNA (TGC PAM) | | Descriptor: | CRISPR-associated endonuclease Cas9/Csn1, non-targeted DNA, sgRNA, ... | | Authors: | Chen, W, Zhang, H, Zhang, Y, Wang, Y, Gan, J, Ji, Q. | | Deposit date: | 2019-05-26 | | Release date: | 2019-09-25 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (3.01 Å) | | Cite: | Molecular basis for the PAM expansion and fidelity enhancement of an evolved Cas9 nuclease.
Plos Biol., 17, 2019
|
|
6K57
 
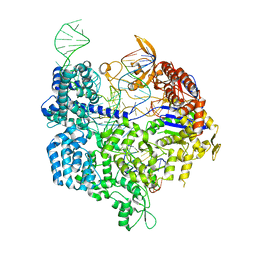 | | Crystal structure of dCas9 in complex with sgRNA and DNA (CGA PAM) | | Descriptor: | CRISPR-associated endonuclease Cas9, non-target DNA, sgRNA, ... | | Authors: | Chen, W, Zhang, H, Zhang, Y, Wang, Y, Gan, J, Ji, Q. | | Deposit date: | 2019-05-28 | | Release date: | 2019-09-25 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (2.98 Å) | | Cite: | Molecular basis for the PAM expansion and fidelity enhancement of an evolved Cas9 nuclease.
Plos Biol., 17, 2019
|
|
5F2V
 
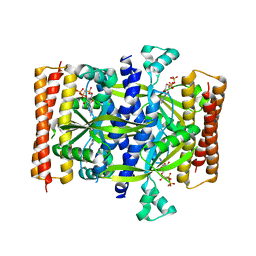 | | Crystal structure of the small alarmone synthethase 1 from Bacillus subtilis bound to AMPCPP | | Descriptor: | DIPHOSPHOMETHYLPHOSPHONIC ACID ADENOSYL ESTER, GTP pyrophosphokinase YjbM, MAGNESIUM ION | | Authors: | Steinchen, W, Schuhmacher, J.S, Altegoer, F, Bange, G. | | Deposit date: | 2015-12-02 | | Release date: | 2015-12-16 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Catalytic mechanism and allosteric regulation of an oligomeric (p)ppGpp synthetase by an alarmone.
Proc.Natl.Acad.Sci.USA, 112, 2015
|
|
5D2Q
 
 | |
5D23
 
 | |
4X36
 
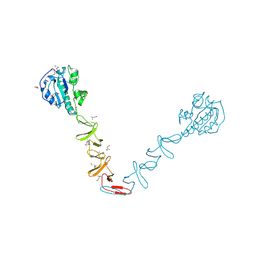 | | Crystal structure of the autolysin LytA from Streptococcus pneumoniae TIGR4 | | Descriptor: | Autolysin, CHOLINE ION, GLYCEROL, ... | | Authors: | Cheng, W, Li, Q, Zhou, C.Z, Chen, Y.X. | | Deposit date: | 2014-11-28 | | Release date: | 2015-05-27 | | Last modified: | 2015-06-24 | | Method: | X-RAY DIFFRACTION (2.101 Å) | | Cite: | Full-length structure of the major autolysin LytA.
Acta Crystallogr.,Sect.D, 71, 2015
|
|
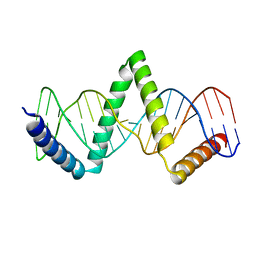
5D2S
 
 | |
4NPL
 
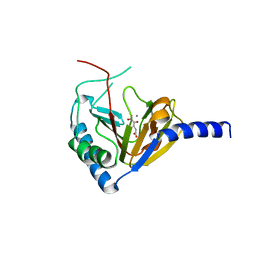 | |
2M9J
 
 | | NMR solution structure of Pin1 WW domain mutant 6-1g | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, Peptidyl-prolyl cis-trans isomerase NIMA-interacting 1 | | Authors: | Enck, S, Chen, W, Price, J.L, Powers, E.T, Wong, C, Dyson, H.J, Kelly, J.W. | | Deposit date: | 2013-06-10 | | Release date: | 2013-06-26 | | Last modified: | 2020-07-29 | | Method: | SOLUTION NMR | | Cite: | Structural and energetic basis of carbohydrate-aromatic packing interactions in proteins.
J.Am.Chem.Soc., 135, 2013
|
|
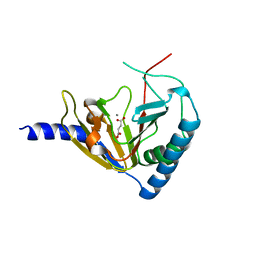
4NPM
 
 | |
2M9F
 
 | | NMR solution structure of Pin1 WW domain mutant 5-1g | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, Peptidyl-prolyl cis-trans isomerase NIMA-interacting 1 | | Authors: | Enck, S, Chen, W, Price, J.L, Powers, E.T, Wong, C, Dyson, H.J, Kelly, J.W. | | Deposit date: | 2013-06-07 | | Release date: | 2013-06-26 | | Last modified: | 2023-06-14 | | Method: | SOLUTION NMR | | Cite: | Structural and energetic basis of carbohydrate-aromatic packing interactions in proteins.
J.Am.Chem.Soc., 135, 2013
|
|
2M9E
 
 | | NMR solution structure of Pin1 WW domain mutant 5-1 | | Descriptor: | Peptidyl-prolyl cis-trans isomerase NIMA-interacting 1 | | Authors: | Enck, S, Chen, W, Price, J.L, Powers, E.T, Wong, C, Dyson, H.J, Kelly, J.W. | | Deposit date: | 2013-06-07 | | Release date: | 2013-06-26 | | Last modified: | 2024-05-15 | | Method: | SOLUTION NMR | | Cite: | Structural and energetic basis of carbohydrate-aromatic packing interactions in proteins.
J.Am.Chem.Soc., 135, 2013
|
|
2M9I
 
 | | NMR solution structure of Pin1 WW domain variant 6-1 | | Descriptor: | Peptidyl-prolyl cis-trans isomerase NIMA-interacting 1 | | Authors: | Enck, S, Chen, W, Price, J.L, Powers, E.T, Wong, C, Dyson, H.J, Kelly, J.W. | | Deposit date: | 2013-06-10 | | Release date: | 2013-06-26 | | Last modified: | 2024-05-15 | | Method: | SOLUTION NMR | | Cite: | Structural and energetic basis of carbohydrate-aromatic packing interactions in proteins.
J.Am.Chem.Soc., 135, 2013
|
|
