7EZT
 
 | | The structure and functional mechanism of nucleotide regulated acetylhexosaminidase Am2136 from Akkermansia muciniphila | | Descriptor: | Beta-N-acetylhexosaminidase, MAGNESIUM ION | | Authors: | Bao, R, Li, C.C, Tang, X.Y, Zhu, Y.B, Song, Y.J, Zhao, N.L, Huang, Q, Mou, X.Y, Luo, G.H, Liu, T.G. | | Deposit date: | 2021-06-02 | | Release date: | 2022-11-02 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (2.81 Å) | | Cite: | Nucleotide binding as an allosteric regulatory mechanism for Akkermansia muciniphila beta- N -acetylhexosaminidase Am2136.
Gut Microbes, 14, 2022
|
|
5WYB
 
 | | Structure of Pseudomonas aeruginosa DspI | | Descriptor: | (4R)-2-METHYLPENTANE-2,4-DIOL, ACETATE ION, Probable enoyl-CoA hydratase/isomerase | | Authors: | Liu, L, Peng, C, Li, T, Li, C, He, L, Song, Y, Zhu, Y, Shen, Y, Bao, R. | | Deposit date: | 2017-01-12 | | Release date: | 2018-01-17 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (2.25 Å) | | Cite: | Structural and functional studies on Pseudomonas aeruginosa DspI: implications for its role in DSF biosynthesis.
Sci Rep, 8, 2018
|
|
6L3X
 
 | | Discovery of novel peptidomimetic boronate ClpP inhibitors with noncanonical enzyme mechanism as potent virulence blockers in vitro and in vivo | | Descriptor: | ATP-dependent Clp protease proteolytic subunit, [(1~{R})-1-[[(2~{S})-2-[[2,5-bis(chloranyl)phenyl]carbonylamino]-3-(1~{H}-indol-3-yl)propanoyl]amino]-3-methyl-butyl]boronic acid | | Authors: | Luo, Y.F, Bao, R, Ju, Y, He, L.H. | | Deposit date: | 2019-10-15 | | Release date: | 2020-08-26 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (2.3054 Å) | | Cite: | Discovery of Novel Peptidomimetic Boronate ClpP Inhibitors with Noncanonical Enzyme Mechanism as Potent Virulence Blockersin Vitroandin Vivo.
J.Med.Chem., 63, 2020
|
|
8Z6G
 
 | |
8YLB
 
 | | Cocrystal structures of agonists compound 1 with HsClpP | | Descriptor: | 5-[(2-methylphenyl)methyl]-11-(phenylmethyl)-2,5,7,11-tetrazatricyclo[7.4.0.0^{2,6}]trideca-1(9),6-dien-8-one, ATP-dependent Clp protease proteolytic subunit, mitochondrial | | Authors: | Zhao, N, Zhu, Y, Bao, R. | | Deposit date: | 2024-03-06 | | Release date: | 2024-05-15 | | Method: | X-RAY DIFFRACTION (2.15 Å) | | Cite: | Rational Design of a Novel Class of Human ClpP Agonists through a Ring-Opening Strategy with Enhanced Antileukemia Activity.
J.Med.Chem., 67, 2024
|
|
5WYD
 
 | | Structural of Pseudomonas aeruginosa DspI | | Descriptor: | (4R)-2-METHYLPENTANE-2,4-DIOL, (4S)-2-METHYL-2,4-PENTANEDIOL, ISOPROPYL ALCOHOL, ... | | Authors: | Liu, L, Peng, C, Li, T, Li, C, He, L, Song, Y, Zhu, Y, Shen, Y, Bao, R. | | Deposit date: | 2017-01-12 | | Release date: | 2018-01-31 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (2.101 Å) | | Cite: | Structural and functional studies on Pseudomonas aeruginosa DspI: implications for its role in DSF biosynthesis.
Sci Rep, 8, 2018
|
|
7BOV
 
 | | The Structure of Bacillus subtilis glycosyltransferase,Bs-YjiC | | Descriptor: | GLYCEROL, SODIUM ION, Uncharacterized UDP-glucosyltransferase YjiC | | Authors: | Zhao, C, Liu, B, Zhao, N.L, Luo, Y.Z, Bao, R. | | Deposit date: | 2020-03-20 | | Release date: | 2020-05-13 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2.293 Å) | | Cite: | Structural and biochemical studies of the glycosyltransferase Bs-YjiC from Bacillus subtilis.
Int.J.Biol.Macromol., 166, 2021
|
|
7BOR
 
 | | Structure of Pseudomonas aeruginosa CoA-bound OdaA | | Descriptor: | COENZYME A, Probable enoyl-CoA hydratase/isomerase | | Authors: | Zhao, N, Zhao, C, Liu, L, Li, T, Li, C, He, L, Zhu, Y, Song, Y, Bao, R. | | Deposit date: | 2020-03-19 | | Release date: | 2020-05-13 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (1.901 Å) | | Cite: | Structural and molecular dynamic studies of Pseudomonas aeruginosa OdaA reveal the regulation role of a C-terminal hinge element.
Biochim Biophys Acta Gen Subj, 1865, 2020
|
|
3B54
 
 | |
8JU7
 
 | |
8KHG
 
 | | Itaconyl-CoA hydratase PaIch | | Descriptor: | CITRIC ACID, Itaconyl-CoA hydratase | | Authors: | Huang, Q, Bao, R. | | Deposit date: | 2023-08-21 | | Release date: | 2024-05-15 | | Last modified: | 2024-07-24 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Structural and functional characterization of itaconyl-CoA hydratase and citramalyl-CoA lyase involved in itaconate metabolism of Pseudomonas aeruginosa.
Structure, 32, 2024
|
|
8KHL
 
 | | (S)-citramalyl-CoA lyase | | Descriptor: | COENZYME A, Probable acyl-CoA lyase beta chain | | Authors: | Huang, Q, Bao, R. | | Deposit date: | 2023-08-22 | | Release date: | 2024-05-15 | | Last modified: | 2024-07-24 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Structural and functional characterization of itaconyl-CoA hydratase and citramalyl-CoA lyase involved in itaconate metabolism of Pseudomonas aeruginosa.
Structure, 32, 2024
|
|
8JX4
 
 | | Structure of the catalytic domain of pseudomurein endo-isopeptidases PeiW | | Descriptor: | ALANINE, D-GLUTAMIC ACID, PeiW | | Authors: | Guo, L.Z, Zhao, N.L, Len, H, Wang, S.X, Cha, G.H, Bai, L.p, Bao, R. | | Deposit date: | 2023-06-30 | | Release date: | 2024-10-09 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Characterization of Structure and Function of Pseudomurein Endoisopeptidases: peiW and peiP
To Be Published
|
|
7XAQ
 
 | | Cryo-EM structure of PvrA-DNA complex | | Descriptor: | Probable transcriptional regulator, fadD1 | | Authors: | Zhu, Y.B, Su, Z.M, Bao, R. | | Deposit date: | 2022-03-18 | | Release date: | 2023-04-26 | | Last modified: | 2024-06-26 | | Method: | ELECTRON MICROSCOPY (3.59 Å) | | Cite: | The Pseudomonas aeruginosa regulator PvrA binds cooperatively to multiple pseudo-palindromic sites to efficiently stimulate target gene
To be published
|
|
8WCO
 
 | | (S)-citramalyl-CoA lyase | | Descriptor: | ACETYL COENZYME *A, Probable acyl-CoA lyase beta chain | | Authors: | Huang, Q, Bao, R. | | Deposit date: | 2023-09-13 | | Release date: | 2024-05-15 | | Last modified: | 2024-07-24 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Structural and functional characterization of itaconyl-CoA hydratase and citramalyl-CoA lyase involved in itaconate metabolism of Pseudomonas aeruginosa.
Structure, 32, 2024
|
|
3PUB
 
 | | Crystal structure of the Bombyx mori low molecular weight lipoprotein 7 (Bmlp7) | | Descriptor: | 30kDa protein | | Authors: | Yang, J.-P, Ma, X.-X, He, Y.-X, Li, W.-F, Kang, Y, Bao, R, Chen, Y, Zhou, C.-Z. | | Deposit date: | 2010-12-03 | | Release date: | 2011-06-01 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (1.91 Å) | | Cite: | Crystal structure of the 30 K protein from the silkworm Bombyx mori reveals a new member of the beta-trefoil superfamily
J.Struct.Biol., 175, 2011
|
|
3EVI
 
 | | Crystal structure of the thioredoxin-fold domain of human phosducin-like protein 2 | | Descriptor: | DI(HYDROXYETHYL)ETHER, Phosducin-like protein 2, SODIUM ION | | Authors: | Lou, X, Bao, R, Zhou, C, Chen, Y. | | Deposit date: | 2008-10-13 | | Release date: | 2009-03-03 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | Structure of the thioredoxin-fold domain of human phosducin-like protein 2
Acta Crystallogr.,Sect.F, 65, 2009
|
|
3F3H
 
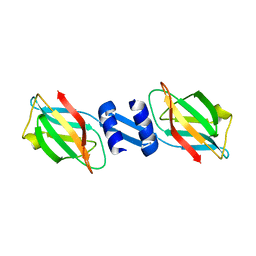 | | Crystal structure and anti-tumor activity of LZ-8 from the fungus Ganoderma lucidium | | Descriptor: | Immunomodulatory protein Ling Zhi-8 | | Authors: | Zhou, C.Z, Huang, L, He, Y.X, Bao, R, Sun, F, Liang, C, Liu, L. | | Deposit date: | 2008-10-30 | | Release date: | 2009-09-15 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Crystal structure of LZ-8 from the medicinal fungus Ganoderma lucidium
Proteins, 75, 2009
|
|
3D8X
 
 | | Crystal Structure of Saccharomyces cerevisiae NDPPH Dependent Thioredoxin Reductase 1 | | Descriptor: | FLAVIN-ADENINE DINUCLEOTIDE, NADPH DIHYDRO-NICOTINAMIDE-ADENINE-DINUCLEOTIDE PHOSPHATE, Thioredoxin reductase 1 | | Authors: | Zhang, Z.Y, Bao, R, Yu, J, Chen, Y.X, Zhou, C.-Z. | | Deposit date: | 2008-05-26 | | Release date: | 2008-12-09 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Crystal structure of Saccharomyces cerevisiae cytoplasmic thioredoxin reductase Trr1 reveals the structural basis for species-specific recognition of thioredoxin
Biochim.Biophys.Acta, 1794, 2009
|
|
6L40
 
 | | Discovery of novel peptidomimetic boronate ClpP inhibitors with noncanonical enzyme mechanism as potent virulence blockers in vitro and in vivo | | Descriptor: | ATP-dependent Clp protease proteolytic subunit, [(1S)-3-methyl-1-[[(2S)-3-phenyl-2-(pyrazin-2-ylcarbonylamino)propanoyl]amino]butyl]boronic acid | | Authors: | Luo, Y.F, Bao, R, Ju, Y, He, L.H. | | Deposit date: | 2019-10-15 | | Release date: | 2020-07-08 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (2.209 Å) | | Cite: | Discovery of Novel Peptidomimetic Boronate ClpP Inhibitors with Noncanonical Enzyme Mechanism as Potent Virulence Blockersin Vitroandin Vivo.
J.Med.Chem., 63, 2020
|
|
5YKJ
 
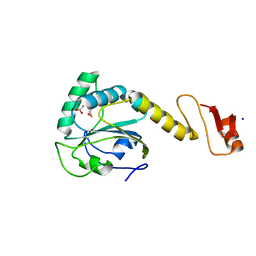 | | Structural basis of the thiol resolving mechanism in yeast mitochondrial 1-Cys peroxiredoxin via glutathione/thioredoxin systems | | Descriptor: | GLYCEROL, Peroxiredoxin PRX1, mitochondrial, ... | | Authors: | Li, C.C, Yang, J, Yang, M.J, Liu, L, Peng, C.T, Li, T, He, L.H, Song, Y.J, Zhu, Y.B, Zhao, N.L, Zhao, C, Bao, R. | | Deposit date: | 2017-10-14 | | Release date: | 2018-10-24 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (1.53 Å) | | Cite: | Structural basis of the thiol resolving mechanism in yeast mitochondrial 1-Cys peroxiredoxin via glutathione/thioredoxin systems
To be published
|
|
5YEI
 
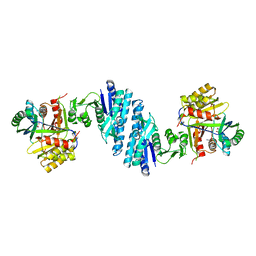 | | Mechanistic insight into the regulation of Pseudomonas aeruginosa aspartate kinase | | Descriptor: | Aspartokinase, GLYCEROL, LYSINE, ... | | Authors: | Li, C, Yang, M, Liu, L, Peng, C, Li, T, He, L, Song, Y, Zhu, Y, Bao, R. | | Deposit date: | 2017-09-17 | | Release date: | 2018-08-29 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (2.301 Å) | | Cite: | Mechanistic insights into the allosteric regulation of Pseudomonas aeruginosa aspartate kinase.
Biochem.J., 475, 2018
|
|
5YKW
 
 | | Structural basis of the thiol resolving mechanism in yeast mitochondrial 1-Cys peroxiredoxin via glutathione/thioredoxin systems | | Descriptor: | Thioredoxin-3, mitochondrial, peptide THR-PRO-VAL-CYS-THR-THR-GLU-VAL | | Authors: | Li, C.C, Yang, J, Yang, M.J, Liu, L, Peng, C.T, Li, T, He, L.H, Song, Y.J, Zhu, Y.B, Zhao, N.L, Zhao, C, Bao, R. | | Deposit date: | 2017-10-16 | | Release date: | 2018-10-24 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (2.08 Å) | | Cite: | Structural basis of the thiol resolving mechanism in yeast mitochondrial 1-Cys peroxiredoxin via glutathione/thioredoxin systems
to be published
|
|
5YLO
 
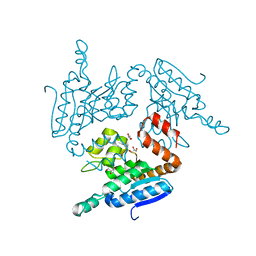 | | Structural of Pseudomonas aeruginosa PA4980 | | Descriptor: | GLYCEROL, Probable enoyl-CoA hydratase/isomerase | | Authors: | Liu, L, Li, T, Peng, C.T, Li, C.C, Xiao, Q.J, He, L.H, Wang, N.Y, Bao, R. | | Deposit date: | 2017-10-18 | | Release date: | 2018-08-22 | | Last modified: | 2024-03-27 | | Method: | X-RAY DIFFRACTION (2.39 Å) | | Cite: | Structural characterization of a Delta3, Delta2-enoyl-CoA isomerase from Pseudomonas aeruginosa: implications for its involvement in unsaturated fatty acid metabolism.
J.Biomol.Struct.Dyn., 37, 2019
|
|
3HIA
 
 | | Crystal structure of the choline binding domain of Spr1274 in Streptococcus pneumoniae | | Descriptor: | Choline binding protein, PHOSPHOCHOLINE, SULFATE ION | | Authors: | Zhang, Z.-Y, Li, W.-Z, Jiang, Y.-L, Bao, R, Zhou, C.-Z, Chen, Y.-X. | | Deposit date: | 2009-05-19 | | Release date: | 2009-08-04 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (2.38 Å) | | Cite: | Crystal structure of the choline binding domain of Spr1274 in Streptococcus pneumoniae
To be Published
|
|
