2YIT
 
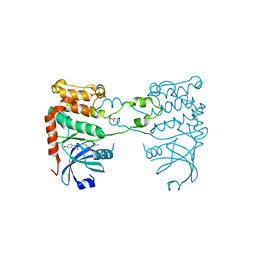 | | Structural analysis of checkpoint kinase 2 in complex with PV1162, a novel inhibitor | | Descriptor: | N-{4-[(1E)-N-carbamimidoylbutanehydrazonoyl]phenyl}-5-methoxy-1H-indole-2-carboxamide, NITRATE ION, SERINE/THREONINE-PROTEIN KINASE CHK2 | | Authors: | Lountos, G.T, Jobson, A.G, Tropea, J.E, Self, C, Zhang, G, Pommier, Y, Shoemaker, R.H, Waugh, D.S. | | Deposit date: | 2011-05-16 | | Release date: | 2011-09-07 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | X-Ray Structures of Checkpoint Kinase 2 in Complex with Inhibitors that Target its Gatekeeper-Dependent Hydrophobic Pocket.
FEBS Lett., 585, 2011
|
|
2YIR
 
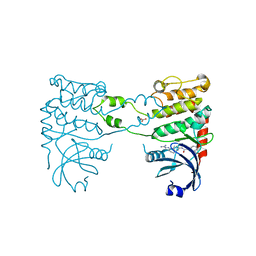 | | Structural analysis of checkpoint kinase 2 in complex with inhibitor PV1352 | | Descriptor: | (E)-N-(5-(2-CARBAMIMIDOYLHYDRAZONO)-5,6,7,8-TETRAHYDRONAPHTHALEN-2-YL)-7-NITRO-1H-INDOLE-2-CARBOXAMIDE, NITRATE ION, SERINE/THREONINE-PROTEIN KINASE CHK2 | | Authors: | Lountos, G.T, Jobson, A.G, Tropea, J.E, Self, C, Zhang, G, Pommier, Y, Shoemaker, R.H, Waugh, D.S. | | Deposit date: | 2011-05-16 | | Release date: | 2011-09-07 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | X-Ray Structures of Checkpoint Kinase 2 in Complex with Inhibitors that Target its Gatekeeper-Dependent Hydrophobic Pocket.
FEBS Lett., 585, 2011
|
|
6MJ5
 
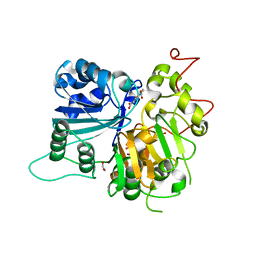 | | Crystal structure of Tdp1 catalytic domain in complex with compound XZ519 | | Descriptor: | 1,2-ETHANEDIOL, 4-hydroxy-8-nitroquinoline-3-carboxylic acid, Tyrosyl-DNA phosphodiesterase 1 | | Authors: | Lountos, G.T, Zhao, X.Z, Kiselev, E, Tropea, J.E, Needle, D, Burke Jr, T.R, Pommier, Y, Waugh, D.S. | | Deposit date: | 2018-09-20 | | Release date: | 2019-07-03 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (1.853 Å) | | Cite: | Identification of a ligand binding hot spot and structural motifs replicating aspects of tyrosyl-DNA phosphodiesterase I (TDP1) phosphoryl recognition by crystallographic fragment cocktail screening.
Nucleic Acids Res., 47, 2019
|
|
6N0O
 
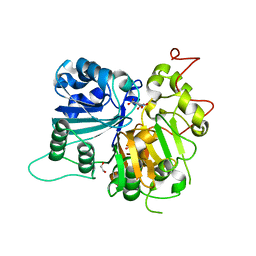 | | Crystal structure of Tdp1 catalytic domain in complex with compound XZ523 | | Descriptor: | 1,2-ETHANEDIOL, 4-nitrobenzene-1,2-dicarboxylic acid, Tyrosyl-DNA phosphodiesterase 1 | | Authors: | Lountos, G.T, Zhao, X.Z, Kiselev, E, Tropea, J.E, Needle, D, Burke Jr, T.R, Pommier, Y, Waugh, D.S. | | Deposit date: | 2018-11-07 | | Release date: | 2019-11-13 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (1.943 Å) | | Cite: | Crystal structure of Tdp1 catalytic domain
To Be Published
|
|
6N19
 
 | | Crystal structure of Tdp1 catalytic domain in complex with compound XZ578 | | Descriptor: | 1,2-ETHANEDIOL, 4-[(4-carboxybutanoyl)amino]benzene-1,2-dicarboxylic acid, Tyrosyl-DNA phosphodiesterase 1 | | Authors: | Lountos, G.T, Zhao, X.Z, Kiselev, E, Tropea, J.E, Needle, D, Burke Jr, T.R, Pommier, Y, Waugh, D.S. | | Deposit date: | 2018-11-08 | | Release date: | 2019-07-03 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (1.501 Å) | | Cite: | Identification of a ligand binding hot spot and structural motifs replicating aspects of tyrosyl-DNA phosphodiesterase I (TDP1) phosphoryl recognition by crystallographic fragment cocktail screening.
Nucleic Acids Res., 47, 2019
|
|
6MYZ
 
 | | Crystal structure of Tdp1 catalytic domain in complex with compound XZ520 | | Descriptor: | 1,2-ETHANEDIOL, 4-oxo-8-phenyl-1,4-dihydroquinoline-3-carboxylic acid, Tyrosyl-DNA phosphodiesterase 1 | | Authors: | Lountos, G.T, Zhao, X.Z, Kiselev, E, Tropea, J.E, Needle, D, Burke Jr, T.R, Pommier, Y, Waugh, D.S. | | Deposit date: | 2018-11-02 | | Release date: | 2019-11-06 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (1.661 Å) | | Cite: | Structure of Tdp1 catalytic domain in complex with compound XZ520
To Be Published
|
|
2IF5
 
 | | Structure of the POZ domain of human LRF, a master regulator of oncogenesis | | Descriptor: | PRASEODYMIUM ION, Zinc finger and BTB domain-containing protein 7A | | Authors: | Schubot, F.D, Waugh, D.S, Tropea, J. | | Deposit date: | 2006-09-20 | | Release date: | 2006-11-21 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Structure of the POZ domain of human LRF, a master regulator of oncogenesis.
Biochem.Biophys.Res.Commun., 351, 2006
|
|
2EZ6
 
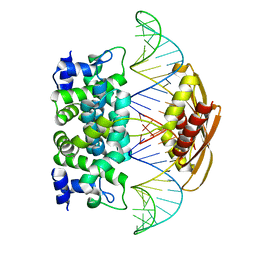 | | Crystal structure of Aquifex aeolicus RNase III (D44N) complexed with product of double-stranded RNA processing | | Descriptor: | 28-MER, MAGNESIUM ION, Ribonuclease III | | Authors: | Gan, J, Tropea, J.E, Austin, B.P, Court, D.L, Waugh, D.S, Ji, X. | | Deposit date: | 2005-11-10 | | Release date: | 2006-02-07 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (2.05 Å) | | Cite: | Structural Insight into the Mechanism of Double-Stranded RNA Processing by Ribonuclease III.
Cell(Cambridge,Mass.), 124, 2006
|
|
2JLH
 
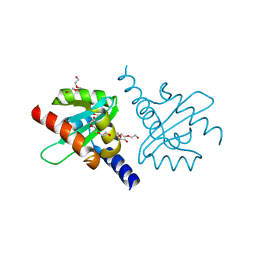 | | Crystal Structure of the Cytoplasmic domain of Yersinia Pestis YscU N263A mutant | | Descriptor: | PENTAETHYLENE GLYCOL, YOP PROTEINS TRANSLOCATION PROTEIN U | | Authors: | Lountos, G.T, Austin, B.P, Nallamsetty, S, Waugh, D.S. | | Deposit date: | 2008-09-09 | | Release date: | 2009-02-03 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (1.53 Å) | | Cite: | Atomic Resolution Structure of the Cytoplasmic Domain of Yersinia Pestis Yscu, a Regulatory Switch Involved in Type III Secretion.
Protein Sci., 18, 2009
|
|
2JR0
 
 | | Solution structure of NusB from Aquifex Aeolicus | | Descriptor: | N utilization substance protein B homolog | | Authors: | Das, R, Loss, S, Li, J, Tarasov, S, Wingfield, P, Waugh, D.S, Byrd, R.A, Altieri, A.S. | | Deposit date: | 2007-06-18 | | Release date: | 2008-02-19 | | Last modified: | 2024-05-08 | | Method: | SOLUTION NMR | | Cite: | Structural biophysics of the NusB:NusE antitermination complex.
J.Mol.Biol., 376, 2008
|
|
1ELJ
 
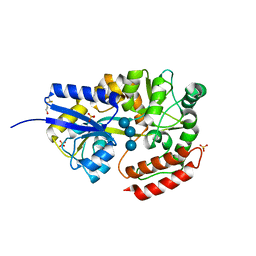 | | THE CRYSTAL STRUCTURE OF LIGANDED MALTODEXTRIN-BINDING PROTEIN FROM PYROCOCCUS FURIOSUS | | Descriptor: | MALTODEXTRIN-BINDING PROTEIN, SULFATE ION, alpha-D-glucopyranose-(1-4)-alpha-D-glucopyranose-(1-4)-alpha-D-glucopyranose | | Authors: | Evdokimov, A.G, Anderson, E.D, Routzahn, K.M, Waugh, D.S. | | Deposit date: | 2000-03-13 | | Release date: | 2001-01-19 | | Last modified: | 2020-07-29 | | Method: | X-RAY DIFFRACTION (1.85 Å) | | Cite: | Structural basis for oligosaccharide recognition by Pyrococcus furiosus maltodextrin-binding protein.
J.Mol.Biol., 305, 2001
|
|
1G9U
 
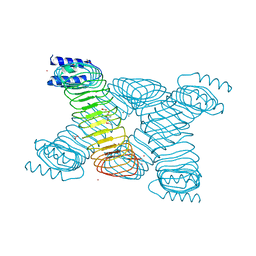 | | CRYSTAL STRUCTURE OF YOPM-LEUCINE RICH EFFECTOR PROTEIN FROM YERSINIA PESTIS | | Descriptor: | ACETATE ION, CALCIUM ION, MERCURY (II) ION, ... | | Authors: | Evdokimov, A.G, Anderson, D.E, Routzahn, K.M, Waugh, D.S. | | Deposit date: | 2000-11-28 | | Release date: | 2001-10-10 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (2.35 Å) | | Cite: | Unusual molecular architecture of the Yersinia pestis cytotoxin YopM: a leucine-rich repeat protein with the shortest repeating unit.
J.Mol.Biol., 312, 2001
|
|
6MNU
 
 | | Crystal structure of Yersinia pestis UDP-glucose pyrophosphorylase | | Descriptor: | 1,2-ETHANEDIOL, UTP--glucose-1-phosphate uridylyltransferase | | Authors: | Gibbs, M.E, Lountos, G.T, Gumpena, R, Waugh, D.S. | | Deposit date: | 2018-10-03 | | Release date: | 2019-09-11 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (2.172 Å) | | Cite: | Crystal structure of UDP-glucose pyrophosphorylase from Yersinia pestis, a potential therapeutic target against plague.
Acta Crystallogr.,Sect.F, 75, 2019
|
|
6MZ0
 
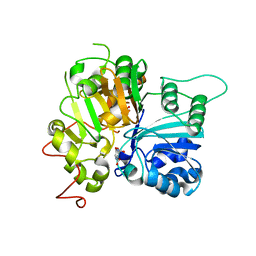 | | Crystal structure of Tdp1 catalytic domain in complex with compound XZ530 | | Descriptor: | 1,2-ETHANEDIOL, 5-hydroxypyrazine-2,3-dicarboxylic acid, Tyrosyl-DNA phosphodiesterase 1 | | Authors: | Lountos, G.T, Zhao, X.Z, Kiselev, E, Tropea, J.E, Needle, D, Burke Jr, T.R, Pommier, Y, Waugh, D.S. | | Deposit date: | 2018-11-02 | | Release date: | 2019-11-06 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (1.969 Å) | | Cite: | Structure of Tdp1 catalytic domain in complex with compound XZ530
To Be Published
|
|
6N17
 
 | | Crystal structure of Tdp1 catalytic domain in complex with compound XZ577 | | Descriptor: | 1,2-ETHANEDIOL, 4-[(3-carboxypropanoyl)amino]benzene-1,2-dicarboxylic acid, DIMETHYL SULFOXIDE, ... | | Authors: | Lountos, G.T, Zhao, X.Z, Kiselev, E, Tropea, J.E, Needle, D, Burke Jr, T.R, Pommier, Y, Waugh, D.S. | | Deposit date: | 2018-11-08 | | Release date: | 2019-07-03 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (1.639 Å) | | Cite: | Identification of a ligand binding hot spot and structural motifs replicating aspects of tyrosyl-DNA phosphodiesterase I (TDP1) phosphoryl recognition by crystallographic fragment cocktail screening.
Nucleic Acids Res., 47, 2019
|
|
6N0R
 
 | | Crystal structure of Tdp1 catalytic domain in complex with compound XZ572 | | Descriptor: | 1,2-ETHANEDIOL, 4-(methylamino)benzene-1,2-dicarboxylic acid, DIMETHYL SULFOXIDE, ... | | Authors: | Lountos, G.T, Zhao, X.Z, Kiselev, E, Tropea, J.E, Needle, D, Burke Jr, T.R, Pommier, Y, Waugh, D.S. | | Deposit date: | 2018-11-07 | | Release date: | 2019-11-13 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (1.544 Å) | | Cite: | Crystal structure of Tdp1 catalytic domain
To Be Published
|
|
6N0D
 
 | | Crystal structure of Tdp1 catalytic domain in complex with compound XZ575 | | Descriptor: | 1,2-ETHANEDIOL, 4-fluorobenzene-1,2-dicarboxylic acid, Tyrosyl-DNA phosphodiesterase 1 | | Authors: | Lountos, G.T, Zhao, X.Z, Kiselev, E, Tropea, J.E, Needle, D, Burke Jr, T.R, Pommier, Y, Waugh, D.S. | | Deposit date: | 2018-11-07 | | Release date: | 2019-11-13 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (1.453 Å) | | Cite: | Crystal structure of Tdp1 catalytic domain in complex with compound XZ575
To Be Published
|
|
6N0N
 
 | | Crystal structure of Tdp1 catalytic domain in complex with compound XZ574 | | Descriptor: | 1,2-ETHANEDIOL, 4-methylbenzene-1,2-dicarboxylic acid, DIMETHYL SULFOXIDE, ... | | Authors: | Lountos, G.T, Zhao, X.Z, Kiselev, E, Tropea, J.E, Needle, D, Burke Jr, T.R, Pommier, Y, Waugh, D.S. | | Deposit date: | 2018-11-07 | | Release date: | 2019-11-13 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (1.477 Å) | | Cite: | Crystal structure of Tdp1 catalytic domain
To Be Published
|
|
5F6D
 
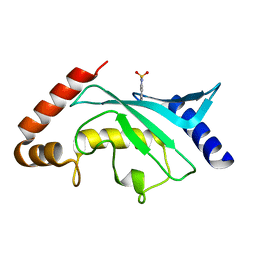 | | Crystal structure of Ubc9 (K48A/K49A/E54A) complexed with Fragment 6 | | Descriptor: | 6~{H}-benzo[c][1,2]benzothiazine 5,5-dioxide, SUMO-conjugating enzyme UBC9 | | Authors: | Lountos, G.T, Hewitt, W.M, Zlotkowski, K, Dahlhauser, S, Saunders, L.B, Needle, D, Tropea, J.E, Zhan, C, Wei, G, Ma, B, Nussinov, R, Schneekloth, J.S.Jr, Waugh, D.S. | | Deposit date: | 2015-12-05 | | Release date: | 2016-04-27 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (1.553 Å) | | Cite: | Insights Into the Allosteric Inhibition of the SUMO E2 Enzyme Ubc9.
Angew.Chem.Int.Ed.Engl., 55, 2016
|
|
5F6W
 
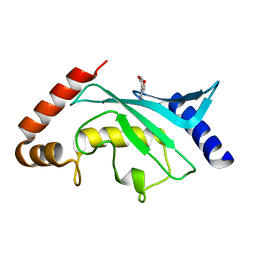 | | Crystal structure of Ubc9 (K48/K49A/E54A) complexed with Fragment 1 (biphenol) | | Descriptor: | 2-(2-hydroxyphenyl)phenol, SUMO-conjugating enzyme UBC9 | | Authors: | Lountos, G.T, Hewitt, W.M, Zlotkowski, K, Dahlhauser, S, Saunders, L.B, Needle, D, Tropea, J.E, Zhan, C, Wei, G, Ma, B, Nussinov, R, Schneekloth, J.S.Jr, Waugh, D.S. | | Deposit date: | 2015-12-07 | | Release date: | 2016-04-27 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (1.699 Å) | | Cite: | Insights Into the Allosteric Inhibition of the SUMO E2 Enzyme Ubc9.
Angew.Chem.Int.Ed.Engl., 55, 2016
|
|
5F6V
 
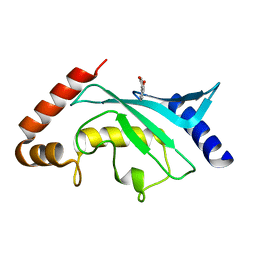 | | Crystal structure of Ubc9 (K48/K49A/E54A) complexed with Fragment 1 (biphenol from fragment cocktail screen) | | Descriptor: | 2-(2-hydroxyphenyl)phenol, SUMO-conjugating enzyme UBC9 | | Authors: | Lountos, G.T, Hewitt, W.M, Zlotkowski, K, Dahlhauser, S, Saunders, L.B, Needle, D, Tropea, J.E, Zhan, C, Wei, G, Ma, B, Nussinov, R, Schneekloth, J.S.Jr, Waugh, D.S. | | Deposit date: | 2015-12-07 | | Release date: | 2016-04-27 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (1.492 Å) | | Cite: | Insights Into the Allosteric Inhibition of the SUMO E2 Enzyme Ubc9.
Angew.Chem.Int.Ed.Engl., 55, 2016
|
|
5F6X
 
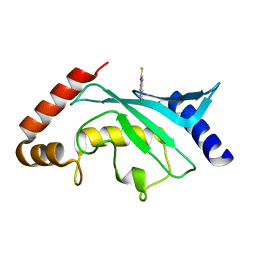 | | Crystal structure of Ubc9 (K48/K49A/E54A) complexed with Fragment 2 (mercaptobenzoxazole from cocktail screen) | | Descriptor: | 5-chloranyl-3~{H}-1,3-benzoxazole-2-thione, SUMO-conjugating enzyme UBC9 | | Authors: | Lountos, G.T, Hewitt, W.M, Zlotkowski, K, Dahlhauser, S, Saunders, L.B, Needle, D, Tropea, J.E, Zhan, C, Wei, G, Ma, B, Nussinov, R, Schneekloth, J.S.Jr, Waugh, D.S. | | Deposit date: | 2015-12-07 | | Release date: | 2016-04-27 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (1.56 Å) | | Cite: | Insights Into the Allosteric Inhibition of the SUMO E2 Enzyme Ubc9.
Angew.Chem.Int.Ed.Engl., 55, 2016
|
|
5F6E
 
 | | Crystal Structure of human Ubc9 (K48A/K49A/E54A) | | Descriptor: | 1,2-ETHANEDIOL, SUMO-conjugating enzyme UBC9 | | Authors: | Lountos, G.T, Hewitt, W.M, Zlotkowski, Z, Dahlhauser, S, Saunders, L.B, Needle, D, Tropea, J.E, Zhan, C, Wei, G, Ma, B, Nussinov, R, Schneekloth, J.S.Jr, Waugh, D.S. | | Deposit date: | 2015-12-05 | | Release date: | 2016-04-27 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (1.12 Å) | | Cite: | Insights Into the Allosteric Inhibition of the SUMO E2 Enzyme Ubc9.
Angew.Chem.Int.Ed.Engl., 55, 2016
|
|
5F6U
 
 | | Crystal Structure of Ubc9 (K48A/K49A/E54A) complexed with Fragment 8 (JSS190B146) | | Descriptor: | SUMO-conjugating enzyme UBC9, ethyl 3-[4-(2-hydroxyphenyl)-3-oxidanyl-phenyl]propanoate | | Authors: | Lountos, G.T, Hewitt, W.M, Zlotkowski, Z, Dahlhauser, S, Saunders, L.B, Needle, D, Tropea, J.E, Zhan, C, Wei, G, Ma, B, Nussinov, R, Schneekloth, J.S.Jr, Waugh, D.S. | | Deposit date: | 2015-12-07 | | Release date: | 2016-04-27 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (1.552 Å) | | Cite: | Insights Into the Allosteric Inhibition of the SUMO E2 Enzyme Ubc9.
Angew.Chem.Int.Ed.Engl., 55, 2016
|
|
5F6Y
 
 | | Crystal structure of Ubc9 (K48/K49A/E54A) complexed with Fragment 2 (mercaptobenzoxazole) | | Descriptor: | 5-chloranyl-3~{H}-1,3-benzoxazole-2-thione, SUMO-conjugating enzyme UBC9 | | Authors: | Lountos, G.T, Hewitt, W.M, Zlotkowski, K, Dahlhauser, S, Saunders, L.B, Needle, D, Tropea, J.E, Zhan, C, Wei, G, Ma, B, Nussinov, R, Schneekloth, J.S.Jr, Waugh, D.S. | | Deposit date: | 2015-12-07 | | Release date: | 2016-04-27 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (1.14 Å) | | Cite: | Insights Into the Allosteric Inhibition of the SUMO E2 Enzyme Ubc9.
Angew.Chem.Int.Ed.Engl., 55, 2016
|
|
