6Y3B
 
 | | Crystal Structure of Unlinked NS2B-NS3 Protease from Zika Virus in Complex with Inhibitor MI-2110 | | Descriptor: | 1-[(8~{R},15~{S},18~{S})-15,18-bis(4-azanylbutyl)-4,7,14,17,20-pentakis(oxidanylidene)-3,6,13,16,19-pentazabicyclo[20.3.1]hexacosa-1(25),22(26),23-trien-8-yl]guanidine, GLYCEROL, Genome polyprotein, ... | | Authors: | Huber, S, Heine, A, Steinmetzer, T. | | Deposit date: | 2020-02-18 | | Release date: | 2020-07-08 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (1.59 Å) | | Cite: | Structure-Based Macrocyclization of Substrate Analogue NS2B-NS3 Protease Inhibitors of Zika, West Nile and Dengue viruses.
Chemmedchem, 15, 2020
|
|
7O2M
 
 | | Crystal Structure of Unlinked NS2B-NS3 Protease from Zika Virus in Complex with Inhibitor MI-2289 | | Descriptor: | 1-[(3~{S},6~{S},9~{S},19~{R})-3,6-bis(4-azanylbutyl)-2,5,8,12,15,18-hexakis(oxidanylidene)-9-(phenylmethyl)-1,4,7,11,14,17-hexazacyclotricos-19-yl]guanidine, Genome polyprotein | | Authors: | Huber, S, Heine, A, Steinmetzer, T. | | Deposit date: | 2021-03-30 | | Release date: | 2022-04-06 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Structure-Based Optimization and Characterization of Macrocyclic Zika Virus NS2B-NS3 Protease Inhibitors.
J.Med.Chem., 65, 2022
|
|
7OC2
 
 | | Crystal Structure of Unlinked NS2B-NS3 Protease from Zika Virus in Complex with Inhibitor MI-2295 | | Descriptor: | Cyclic 1[2-CHLORO-4-METHOXY-PHENYL-OXYMETHYL]-4-[2,6-DICHLORO-PHENYL-OXYMETHYL]-BENZENE-(7-3)-7-BENZYL-1,3-DIMETHYL-8-PIPERAZIN-1-YL-3,7-DIHYDRO-PURINE-2,6-DIONE-(7-19)-N-ACETYL-L-CYSTEINE-(8-25)-[3R-[3A,4A,5B(S*)]]-5-(1-CARBOXY-1-PHOSPHONOETHOXY)-4-HYDROXY-3-(PHOSPHONOOXY)-1-CYCLOHEXENE-1-CARBOXYLIC ACID-()-(6E,11E)-HEPTADECA-6,11-DIENE-9,9-DIYLBIS(PHOSPHONIC ACID), Serine protease NS3, Serine protease subunit NS2B | | Authors: | Huber, S, Heine, A, Steinmetzer, T. | | Deposit date: | 2021-04-25 | | Release date: | 2022-04-06 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | Structure-Based Optimization and Characterization of Macrocyclic Zika Virus NS2B-NS3 Protease Inhibitors.
J.Med.Chem., 65, 2022
|
|
7O55
 
 | |
7OBV
 
 | |
1RMO
 
 | | Probing the Role of Tryptophans in Aequorea Victoria Green Fluorescent Proteins with an Expanded Genetic Code | | Descriptor: | wunen-nonfunctional GFP fusion protein | | Authors: | Budisa, N, Pal, P.P, Alefelder, S, Birle, P, Krywcun, T, Rubini, M, Wenger, W, Bae, J.H, Steiner, T. | | Deposit date: | 2003-11-28 | | Release date: | 2004-06-08 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Probing the role of tryptophans in Aequorea victoria green fluorescent proteins with an expanded genetic code
Biol.Chem., 385, 2004
|
|
1RMM
 
 | | Probing the Role of Tryptophans in Aequorea Victoria Green Fluorescent Proteins with an Expanded Genetic Code | | Descriptor: | SIGF1-GFP fusion protein | | Authors: | Budisa, N, Pal, P.P, Alefelder, S, Birle, P, Krywcun, T, Rubini, M, Wenger, W, Bae, J.H, Steiner, T. | | Deposit date: | 2003-11-28 | | Release date: | 2004-06-08 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Probing the role of tryptophans in Aequorea victoria green fluorescent proteins with an expanded genetic code
Biol.Chem., 385, 2004
|
|
6T9U
 
 | | Bovine Trypsine in complex with the synthetic inhibitor (S)-3'-(N-(1-(4-(3-(tert-butyl)ureido)piperidin-1-yl)-3-(3-carbamimidoylphenyl)-1-oxopropan-2-yl)sulfamoyl)-[1,1'-biphenyl]-3-carboximidamide (MI-490) | | Descriptor: | 1-~{tert}-butyl-3-[1-[(2~{S})-3-(3-carbamimidoylphenyl)-2-[[3-(3-carbamimidoylphenyl)phenyl]sulfonylamino]propanoyl]piperidin-4-yl]urea, CALCIUM ION, Cationic Trypsin, ... | | Authors: | Mueller, J.M, Merkl, S, Keils, A, Pilgram, O, Steinmetzer, T. | | Deposit date: | 2019-10-28 | | Release date: | 2020-11-18 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (1.067509 Å) | | Cite: | Improving the selectivity of 3-amidinophenylalanine-derived matriptase inhibitors
Eur.J.Med.Chem., 2022
|
|
6T89
 
 | | Thrombin in complex with (S)-N-(tert-butyl)-4-(3-(3-carbamimidoylphenyl)-2-((2',4'-dimethoxy-[1,1'-biphenyl])-3-sulfonamido)propanoyl)piperazine-1-carboxamide (MI-498) | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 4-[(2~{S})-3-(3-carbamimidoylphenyl)-2-[[3-(4-methoxy-2-oxidanyl-phenyl)phenyl]sulfonylamino]propanoyl]-~{N}-methyl-piperazine-1-carboxamide, DIMETHYL SULFOXIDE, ... | | Authors: | Ngaha, S.A, Sandner, A, Huber, S, Heine, A, Steinmetzer, T, Pilgram, O. | | Deposit date: | 2019-10-24 | | Release date: | 2020-11-18 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Improving the selectivity of 3-amidinophenylalanine-derived matriptase inhibitors
Eur.J.Med.Chem., 2022
|
|
6T9V
 
 | | Bovine Trypsin in complex with the synthetic inhibitor (S)-3-(3-(4-(3-(tert-butyl)ureido)piperidin-1-yl)-2-((3'-fluoro-4'-(hydroxymethyl)-[1,1'-biphenyl])-3-sulfonamido)-3-oxopropyl)benzimidamide (MI-1904) | | Descriptor: | 1-~{tert}-butyl-3-[1-[(2~{S})-3-(3-carbamimidoylphenyl)-2-[[3-[3-fluoranyl-4-(hydroxymethyl)phenyl]phenyl]sulfonylamino ]propanoyl]piperidin-4-yl]urea, CALCIUM ION, Cationic Trypsin, ... | | Authors: | Merkl, S, Keils, A, Mueller, J.M, Pilgram, O, Steinmetzer, T. | | Deposit date: | 2019-10-28 | | Release date: | 2020-11-18 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (1.12642729 Å) | | Cite: | Improving the selectivity of 3-amidinophenylalanine-derived matriptase inhibitors
Eur.J.Med.Chem., 2022
|
|
6T8A
 
 | | Thrombin in complex with diphenyl ((4-carbamimidoylphenyl)((S)-1-((R)-3-cyclohexyl 2-((phenylmethyl)sulfonamido)propanoyl)pyrrolidine-2-carboxamido)methyl)phosphonate (MI-492) | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, DIMETHYL SULFOXIDE, Hirudin variant-2, ... | | Authors: | Ngaha, S.A, Sandner, A, Huber, S, Heine, A, Steinmetzer, T, Pilgram, O. | | Deposit date: | 2019-10-24 | | Release date: | 2020-11-18 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (1.62 Å) | | Cite: | Thrombin in complex with diphenyl ((4-carbamimidoylphenyl)((S)-1-((R)-3-cyclohexyl-2-((phenylmethyl)sulfonamido)propanoyl)pyrrolidine-2-carboxamido)methyl)phosphonate (MI-492)
to be published
|
|
1RM9
 
 | | Probing the Role of Tryptophans in Aequorea Victoria Green Fluorescent Proteins with an Expanded Genetic Code | | Descriptor: | avermectin-sensitive chloride channel GluCl beta/cyan fluorescent protein fusion | | Authors: | Budisa, N, Pal, P.P, Alefelder, S, Birle, P, Krywcun, T, Rubini, M, Wenger, W, Bae, J.H, Steiner, T. | | Deposit date: | 2003-11-27 | | Release date: | 2004-06-08 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (2.9 Å) | | Cite: | Probing the role of tryptophans in Aequorea victoria green fluorescent proteins with an expanded genetic code
Biol.Chem., 385, 2004
|
|
1RMP
 
 | | Probing the Role of Tryptophans in Aequorea Victoria Green Fluorescent Proteins with an Expanded Genetic Code | | Descriptor: | SIGF1-GFP fusion protein | | Authors: | Budisa, N, Pal, P.P, Alefelder, S, Birle, P, Krywcun, T, Rubini, M, Wenger, W, Bae, J.H, Steiner, T. | | Deposit date: | 2003-11-28 | | Release date: | 2004-06-08 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (3 Å) | | Cite: | Probing the role of tryptophans in Aequorea victoria green fluorescent proteins with an expanded genetic code
Biol.Chem., 385, 2004
|
|
6T9T
 
 | | Matriptase in complex with the synthetic inhibitor (S)-3-(3-(4-(3-(tert-butyl)ureido)piperidin-1-yl)-2-((3'-fluoro-4'-(hydroxymethyl)-[1,1'-biphenyl])-3-sulfonamido)-3-oxopropyl)benzimidamide (MI-1904) | | Descriptor: | 1-~{tert}-butyl-3-[1-[(2~{S})-3-(3-carbamimidoylphenyl)-2-[[3-[3-fluoranyl-4-(hydroxymethyl)phenyl]phenyl]sulfonylamino ]propanoyl]piperidin-4-yl]urea, CHLORIDE ION, Suppressor of tumorigenicity 14 protein | | Authors: | Mueller, J.M, Merkl, S, Keils, A, Pilgram, O, Steinmetzer, T. | | Deposit date: | 2019-10-28 | | Release date: | 2020-11-18 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (1.69 Å) | | Cite: | Improving the selectivity of 3-amidinophenylalanine-derived matriptase inhibitors
Eur.J.Med.Chem., 2022
|
|
4MTB
 
 | |
1EB1
 
 | | Complex structure of human thrombin with N-methyl-arginine inhibitor | | Descriptor: | 3-CYCLOHEXYL-D-ALANYL-L-PROLYL-N~2~-METHYL-L-ARGININE, PEPTIDE INHIBITOR, THROMBIN HEAVY CHAIN, ... | | Authors: | Friedrich, R, Steinmetzer, T, Bode, W. | | Deposit date: | 2001-07-18 | | Release date: | 2002-01-28 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | The Methyl Group of N(Alpha)(Me)Arg-Containing Peptides Disturbs the Active-Site Geometry of Thrombin, Impairing Efficient Cleavage
J.Mol.Biol., 316, 2002
|
|
5JXH
 
 | | Structure the proprotein convertase furin in complex with meta-guanidinomethyl-Phac-RVR-Amba at 2.0 Angstrom resolution. | | Descriptor: | 2UC-ARG-VAL-ARG-00S, CALCIUM ION, CHLORIDE ION, ... | | Authors: | Dahms, S.O, Arciniega, M, Steinmetzer, T, Huber, R, Than, M.E. | | Deposit date: | 2016-05-13 | | Release date: | 2016-10-05 | | Last modified: | 2024-07-10 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Structure of the unliganded form of the proprotein convertase furin suggests activation by a substrate-induced mechanism.
Proc.Natl.Acad.Sci.USA, 113, 2016
|
|
5JXG
 
 | | Structure of the unliganded form of the proprotein convertase furin. | | Descriptor: | CALCIUM ION, CHLORIDE ION, Furin, ... | | Authors: | Dahms, S.O, Arciniega, M, Steinmetzer, T, Huber, R, Than, M.E. | | Deposit date: | 2016-05-13 | | Release date: | 2016-10-05 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Structure of the unliganded form of the proprotein convertase furin suggests activation by a substrate-induced mechanism.
Proc.Natl.Acad.Sci.USA, 113, 2016
|
|
5JXJ
 
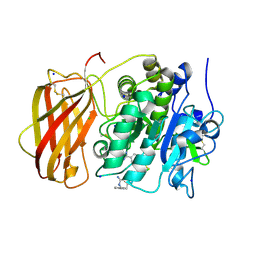 | | Structure of the proprotein convertase furin complexed to meta-guanidinomethyl-Phac-RVR-Amba in presence of EDTA | | Descriptor: | 2UC-ARG-VAL-ARG-00S, CALCIUM ION, CHLORIDE ION, ... | | Authors: | Dahms, S.O, Arciniega, M, Steinmetzer, T, Huber, R, Than, M.E. | | Deposit date: | 2016-05-13 | | Release date: | 2016-10-05 | | Last modified: | 2024-07-10 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Structure of the unliganded form of the proprotein convertase furin suggests activation by a substrate-induced mechanism.
Proc.Natl.Acad.Sci.USA, 113, 2016
|
|
5JXI
 
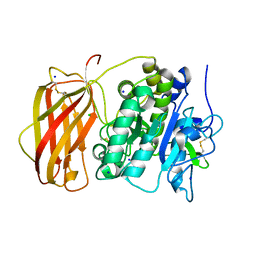 | | Structure of the unliganded form of the proprotein convertase furin in presence of EDTA. | | Descriptor: | CALCIUM ION, CHLORIDE ION, Furin, ... | | Authors: | Dahms, S.O, Arciniega, M, Steinmetzer, T, Huber, R, Than, M.E. | | Deposit date: | 2016-05-13 | | Release date: | 2016-10-05 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Structure of the unliganded form of the proprotein convertase furin suggests activation by a substrate-induced mechanism.
Proc.Natl.Acad.Sci.USA, 113, 2016
|
|
7ZNO
 
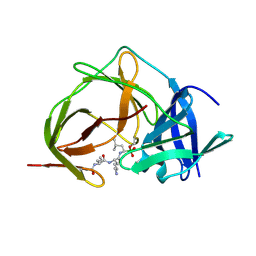 | |
7ZYS
 
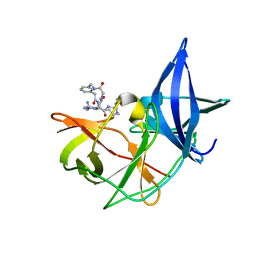 | | Crystal Structure of Unlinked NS2B-NS3 Protease from Zika Virus in Complex with Inhibitor MI-2227 | | Descriptor: | 1-[(8~{R},15~{S},18~{S})-18-(4-azanylbutyl)-15-butyl-4,7,14,17,20-pentakis(oxidanylidene)-3,6,13,16,19-pentazabicyclo[20.3.1]hexacosa-1(26),22,24-trien-8-yl]guanidine, Serine protease NS3, Serine protease subunit NS2B | | Authors: | Huber, S, Steinmetzer, T. | | Deposit date: | 2022-05-25 | | Release date: | 2022-07-13 | | Last modified: | 2024-06-12 | | Method: | X-RAY DIFFRACTION (1.26 Å) | | Cite: | Synthesis and structural characterization of new macrocyclic inhibitors of the Zika virus NS2B-NS3 protease.
Arch Pharm, 2024
|
|
7ZLD
 
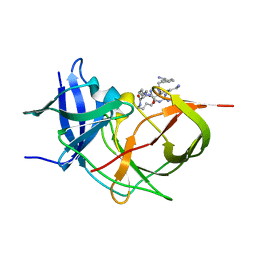 | | Crystal Structure of Unlinked NS2B_NS3 Protease from Zika Virus in Complex with Inhibitor MI-2223 | | Descriptor: | (2~{S})-2-[2-[3-(aminomethyl)phenyl]ethanoylamino]-6-azanyl-~{N}-[(2~{S})-6-azanyl-1-[[(5~{R})-6-azanyl-5-carbamimidamido-6-oxidanylidene-hexyl]amino]-1-oxidanylidene-hexan-2-yl]hexanamide, Serine protease NS3, Serine protease subunit NS2B | | Authors: | Huber, S, Steinmetzer, T. | | Deposit date: | 2022-04-14 | | Release date: | 2022-12-28 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (1.61 Å) | | Cite: | Thermodynamic characterization of a macrocyclic Zika virus NS2B/NS3 protease inhibitor and its acyclic analogs.
Arch Pharm, 356, 2023
|
|
7ZLC
 
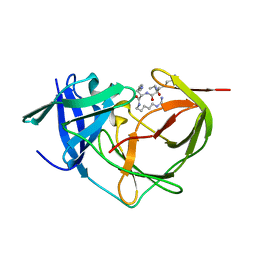 | |
7ZMI
 
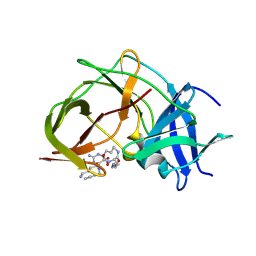 | |
