9IV2
 
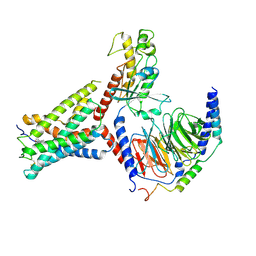 | | Identification, structure and agonist design of an androgen membrane receptor. | | Descriptor: | 5-ALPHA-DIHYDROTESTOSTERONE, Adhesion G-protein coupled receptor D1, Gs protein alpha subunit, ... | | Authors: | Ping, Y.Q, Yang, Z. | | Deposit date: | 2024-07-22 | | Release date: | 2025-02-12 | | Last modified: | 2025-07-02 | | Method: | ELECTRON MICROSCOPY (3.53 Å) | | Cite: | Identification, structure, and agonist design of an androgen membrane receptor.
Cell, 188, 2025
|
|
9IV1
 
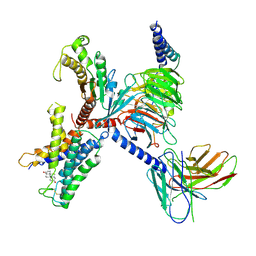 | | Identification, structure and agonist design of an androgen membrane receptor. | | Descriptor: | 5-ALPHA-DIHYDROTESTOSTERONE, Adhesion G-protein coupled receptor D1, Guanine nucleotide-binding protein G(I)/G(S)/G(O) subunit gamma-2, ... | | Authors: | Ping, Y.Q, Yang, Z. | | Deposit date: | 2024-07-22 | | Release date: | 2025-02-12 | | Last modified: | 2025-06-25 | | Method: | ELECTRON MICROSCOPY (2.98 Å) | | Cite: | Identification, structure, and agonist design of an androgen membrane receptor.
Cell, 188, 2025
|
|
7X8R
 
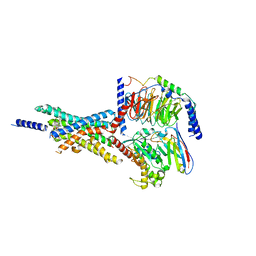 | | Cryo-EM structure of the Boc5-bound hGLP-1R-Gs complex | | Descriptor: | 2,4-bis(3-methoxy-4-thiophen-2-ylcarbonyloxy-phenyl)-1,3-bis[[4-[(2-methylpropan-2-yl)oxycarbonylamino]phenyl]carbonylamino]cyclobutane-1,3-dicarboxylic acid, Glucagon-like peptide 1 receptor, Guanine nucleotide-binding protein G(I)/G(S)/G(O) subunit gamma-2, ... | | Authors: | Cong, Z.T, Zhou, Q.T, Li, Y, Chen, L.N, Zhang, Z.C, Liang, A.Y, Liu, Q, Wu, X.Y, Dai, A.T, Xia, T, Wu, W, Zhang, Y, Yang, D.H, Wang, M.W. | | Deposit date: | 2022-03-14 | | Release date: | 2022-06-29 | | Last modified: | 2024-10-09 | | Method: | ELECTRON MICROSCOPY (2.61 Å) | | Cite: | Structural basis of peptidomimetic agonism revealed by small- molecule GLP-1R agonists Boc5 and WB4-24.
Proc.Natl.Acad.Sci.USA, 119, 2022
|
|
7X8S
 
 | | Cryo-EM structure of the WB4-24-bound hGLP-1R-Gs complex | | Descriptor: | 2,4-bis(3-methoxy-4-thiophen-2-ylcarbonyloxy-phenyl)-1,3-bis[[4-(2-methylpropanoylamino)phenyl]carbonylamino]cyclobutane-1,3-dicarboxylic acid, Glucagon-like peptide 1 receptor, Guanine nucleotide-binding protein G(I)/G(S)/G(O) subunit gamma-2, ... | | Authors: | Cong, Z.T, Zhou, Q.T, Li, Y, Chen, L.N, Zhang, Z.C, Liang, A.Y, Liu, Q, Wu, X.Y, Dai, A.T, Xia, T, Wu, W, Zhang, Y, Yang, D.H, Wang, M.W. | | Deposit date: | 2022-03-14 | | Release date: | 2022-06-29 | | Last modified: | 2024-10-16 | | Method: | ELECTRON MICROSCOPY (3.09 Å) | | Cite: | Structural basis of peptidomimetic agonism revealed by small- molecule GLP-1R agonists Boc5 and WB4-24.
Proc.Natl.Acad.Sci.USA, 119, 2022
|
|
8ZWA
 
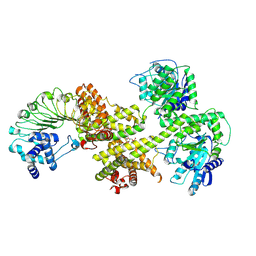 | | HopBY induced At EDS1-PAD4-ADR1 heterotrimer | | Descriptor: | 2'-O-[(5'-PHOSPHO)RIBOSYL]ADENOSINE-5'-MONOPHOSPHATE, Disease resistance protein ADR1, Isoform 1 of Protein EDS1, ... | | Authors: | Xu, W.Y, Zhang, Y, Yu, H, Wan, L. | | Deposit date: | 2024-06-13 | | Release date: | 2024-11-06 | | Last modified: | 2025-01-01 | | Method: | ELECTRON MICROSCOPY (3.48 Å) | | Cite: | Activation of a helper NLR by plant and bacterial TIR immune signaling.
Science, 386, 2024
|
|
8ZW9
 
 | | RPS4-TIR induced At EDS1-PAD4-ADR1 heterotrimer | | Descriptor: | Disease resistance protein ADR1, Isoform 1 of Protein EDS1, PAD4, ... | | Authors: | Xu, W.Y, Zhang, Y, Yu, H, Wan, L. | | Deposit date: | 2024-06-12 | | Release date: | 2024-11-06 | | Last modified: | 2025-01-01 | | Method: | ELECTRON MICROSCOPY (3.03 Å) | | Cite: | Activation of a helper NLR by plant and bacterial TIR immune signaling.
Science, 386, 2024
|
|
8ZB0
 
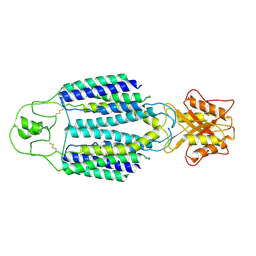 | | Cryo-EM structure of human ZnT1 | | Descriptor: | Proton-coupled zinc antiporter SLC30A1, ZINC ION | | Authors: | Sun, S, Xie, E, Xu, S, Ji, S. | | Deposit date: | 2024-04-25 | | Release date: | 2025-03-12 | | Method: | ELECTRON MICROSCOPY (3.7 Å) | | Cite: | The Intestinal Transporter SLC30A1 Plays a Critical Role in Regulating Systemic Zinc Homeostasis.
Adv Sci, 11, 2024
|
|
2JKU
 
 | | Crystal structure of the N-terminal region of the biotin acceptor domain of human propionyl-CoA carboxylase | | Descriptor: | PROPIONYL-COA CARBOXYLASE ALPHA CHAIN, MITOCHONDRIAL, TETRAETHYLENE GLYCOL | | Authors: | Healy, S, Yue, W.W, Kochan, G, Pilka, E.S, Murray, J.W, Roos, A.K, Filippakopoulos, P, von Delft, F, Arrowsmith, C, Wikstrom, M, Edwards, A, Bountra, C, Gravel, R.A, Oppermann, U. | | Deposit date: | 2008-08-30 | | Release date: | 2008-09-09 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | Structural impact of human and Escherichia coli biotin carboxyl carrier proteins on biotin attachment.
Biochemistry, 49, 2010
|
|
2KLQ
 
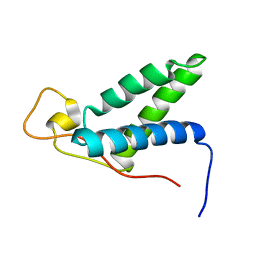 | | The solution structure of CBD of human MCM6 | | Descriptor: | DNA replication licensing factor MCM6 | | Authors: | Liu, C. | | Deposit date: | 2009-07-08 | | Release date: | 2010-03-02 | | Last modified: | 2024-05-29 | | Method: | SOLUTION NMR | | Cite: | Characterization and structure determination of the Cdt1 binding domain of human minichromosome maintenance (Mcm) 6
J.Biol.Chem., 285, 2010
|
|
8ZP2
 
 | |
8ZP1
 
 | |
8ZOY
 
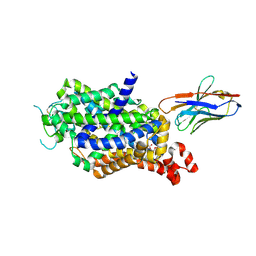 | |
8ZPB
 
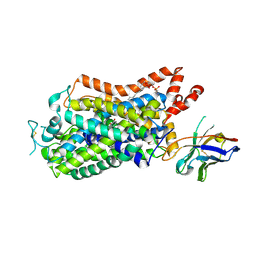 | |
6IUP
 
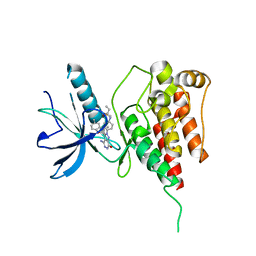 | | Crystal structure of FGFR4 kinase domain in complex with compound 5 | | Descriptor: | DIMETHYL SULFOXIDE, Fibroblast growth factor receptor 4, N-{4-[4-amino-3-(3,5-dimethyl-1-benzofuran-2-yl)-7-oxo-6,7-dihydro-2H-pyrazolo[3,4-d]pyridazin-2-yl]phenyl}prop-2-enamide | | Authors: | Xu, Y, Liu, Q. | | Deposit date: | 2018-11-29 | | Release date: | 2019-11-06 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Discovery and Development of a Series of Pyrazolo[3,4-d]pyridazinone Compounds as the Novel Covalent Fibroblast Growth Factor Receptor Inhibitors by the Rational Drug Design.
J.Med.Chem., 62, 2019
|
|
4HPZ
 
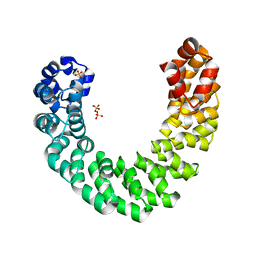 | |
4QBS
 
 | |
9HVW
 
 | |
4I54
 
 | |
4I3S
 
 | | Crystal structure of the outer domain of HIV-1 gp120 in complex with VRC-PG04 space group P21 | | Descriptor: | CALCIUM ION, Heavy chain of VRC-PG04 Fab, Light chain of VRC-PG04 Fab, ... | | Authors: | Joyce, M.G, Biertumpfel, C, Nabel, G.J, Kwong, P.D. | | Deposit date: | 2012-11-26 | | Release date: | 2013-01-09 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (2.85 Å) | | Cite: | Outer Domain of HIV-1 gp120: Antigenic Optimization, Structural Malleability, and Crystal Structure with Antibody VRC-PG04.
J.Virol., 87, 2013
|
|
4QBR
 
 | |
4I53
 
 | |
4R4F
 
 | | Crystal structure of non-neutralizing, A32-like antibody 2.2c in complex with HIV-1 YU2 gp120 | | Descriptor: | (R,R)-2,3-BUTANEDIOL, 2-acetamido-2-deoxy-beta-D-glucopyranose, Antibody 2.2c LIGHT CHAIN, ... | | Authors: | Acharya, P, Kwong, P.D. | | Deposit date: | 2014-08-19 | | Release date: | 2014-09-17 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (3.514 Å) | | Cite: | Structural Definition of an Antibody-Dependent Cellular Cytotoxicity Response Implicated in Reduced Risk for HIV-1 Infection.
J.Virol., 88, 2014
|
|
4H8W
 
 | | Crystal structure of non-neutralizing and ADCC-potent antibody N5-i5 in complex with HIV-1 clade A/E gp120 and sCD4. | | Descriptor: | (4R)-2-METHYLPENTANE-2,4-DIOL, 2-acetamido-2-deoxy-beta-D-glucopyranose, FAB HEAVY CHAIN OF ADCC AND NON-NEUTRALIZING ANTI-HIV-1 ANTIBODY N5-I5, ... | | Authors: | Tolbert, W.D, Acharya, P, Kwong, P.D, Pazgier, M. | | Deposit date: | 2012-09-24 | | Release date: | 2014-04-09 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (1.85 Å) | | Cite: | Structural Definition of an Antibody-Dependent Cellular Cytotoxicity Response Implicated in Reduced Risk for HIV-1 Infection.
J.Virol., 88, 2014
|
|
7V3E
 
 | | The complex structure of soBcmB and its intermediate product 1a | | Descriptor: | (3S,6Z)-3-[(2S)-butan-2-yl]-6-[(2R)-2-methyl-2,3-bis(oxidanyl)propylidene]piperazine-2,5-dione, 2-OXOGLUTARIC ACID, CHLORIDE ION, ... | | Authors: | Wu, L, Zhou, J.H. | | Deposit date: | 2021-08-10 | | Release date: | 2023-02-15 | | Last modified: | 2024-09-04 | | Method: | X-RAY DIFFRACTION (2.12 Å) | | Cite: | Enzymatic catalysis favours eight-membered over five-membered ring closure in bicyclomycin biosynthesis
Nat Catal, 6, 2023
|
|
7V2T
 
 | | The complex structure of SoBcmB and its natural precursor 2 | | Descriptor: | (3S,6S)-3-((R)-2,3-dihydroxy-2-methylpropyl)-6-((S)-4-hydroxybutan-2-yl)piperazine-2,5-dione, 2-OXOGLUTARIC ACID, CHLORIDE ION, ... | | Authors: | Zhou, J.H, Wu, L. | | Deposit date: | 2021-08-09 | | Release date: | 2023-02-15 | | Last modified: | 2024-09-04 | | Method: | X-RAY DIFFRACTION (2.20005679 Å) | | Cite: | Enzymatic catalysis favours eight-membered over five-membered ring closure in bicyclomycin biosynthesis
Nat Catal, 6, 2023
|
|
