6YXK
 
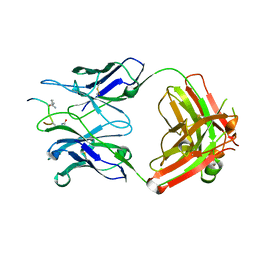 | | Crystal structure of ACPA 3F3 in complex with cit-vimentin 59-74 | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, ACPA 3F3 Fab fragment - heavy chain, ACPA 3F3 Fab fragment - light chain, ... | | Authors: | Ge, C, Holmdahl, R. | | Deposit date: | 2020-05-03 | | Release date: | 2021-05-12 | | Last modified: | 2024-11-20 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Surface Ig variable domain glycosylation affects autoantigen binding and acts as threshold for human autoreactive B cell activation.
Sci Adv, 8, 2022
|
|
6YXL
 
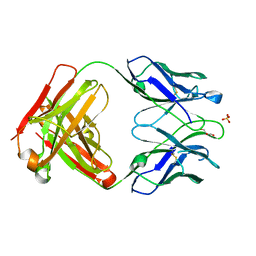 | | Crystal structure of ACPA F3 | | Descriptor: | ACPA F3 Fab fragment - heavy chain, ACPA F3 Fab fragment - light chain, GLYCEROL, ... | | Authors: | Ge, C, Holmdahl, R. | | Deposit date: | 2020-05-03 | | Release date: | 2021-05-12 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Surface Ig variable domain glycosylation affects autoantigen binding and acts as threshold for human autoreactive B cell activation.
Sci Adv, 8, 2022
|
|
6W6Z
 
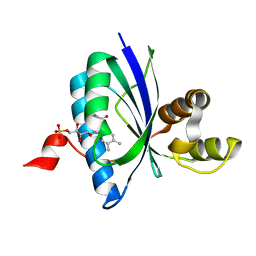 | | BlsA ground state | | Descriptor: | BLUF domain-containing protein, FLAVIN MONONUCLEOTIDE | | Authors: | Chitrakar, I, French, J.B. | | Deposit date: | 2020-03-18 | | Release date: | 2020-10-07 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (1.71 Å) | | Cite: | Structural Basis for the Regulation of Biofilm Formation and Iron Uptake in A. baumannii by the Blue-Light-Using Photoreceptor, BlsA.
Acs Infect Dis., 6, 2020
|
|
6W72
 
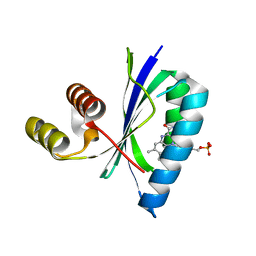 | | BlsA photo-activated state | | Descriptor: | BLUF domain-containing protein, FLAVIN MONONUCLEOTIDE | | Authors: | Chitrakar, I, French, J.B. | | Deposit date: | 2020-03-18 | | Release date: | 2020-10-07 | | Last modified: | 2024-03-06 | | Method: | X-RAY DIFFRACTION (1.76 Å) | | Cite: | Structural Basis for the Regulation of Biofilm Formation and Iron Uptake in A. baumannii by the Blue-Light-Using Photoreceptor, BlsA.
Acs Infect Dis., 6, 2020
|
|
7S4J
 
 | | CryoEM structure of Methylococcus capsulatus (Bath) pMMO in a native lipid nanodisc at 2.16 Angstrom resolution | | Descriptor: | 1,2-DIDECANOYL-SN-GLYCERO-3-PHOSPHOCHOLINE, 1,2-dihexanoyl-sn-glycero-3-phosphocholine, Ammonia monooxygenase/methane monooxygenase, ... | | Authors: | Koo, C.W, Rosenzweig, A.C. | | Deposit date: | 2021-09-09 | | Release date: | 2022-03-30 | | Last modified: | 2024-06-05 | | Method: | ELECTRON MICROSCOPY (2.16 Å) | | Cite: | Recovery of particulate methane monooxygenase structure and activity in a lipid bilayer.
Science, 375, 2022
|
|
7S4H
 
 | | CryoEM structure of Methylococcus capsulatus (Bath) pMMO in a native lipid nanodisc at 2.14 Angstrom resolution | | Descriptor: | 1,2-DIDECANOYL-SN-GLYCERO-3-PHOSPHOCHOLINE, 1,2-dihexanoyl-sn-glycero-3-phosphocholine, Ammonia monooxygenase/methane monooxygenase, ... | | Authors: | Koo, C.W, Rosenzweig, A.C. | | Deposit date: | 2021-09-08 | | Release date: | 2022-03-30 | | Last modified: | 2024-06-05 | | Method: | ELECTRON MICROSCOPY (2.14 Å) | | Cite: | Recovery of particulate methane monooxygenase structure and activity in a lipid bilayer.
Science, 375, 2022
|
|
7S4M
 
 | |
4XAI
 
 | | Crystal Structure of red flour beetle NR2E1/TLX | | Descriptor: | Grunge, isoform J, Maltose-binding periplasmic protein,Tailless ortholog, ... | | Authors: | Zhi, X, Zhou, E, Xu, E. | | Deposit date: | 2014-12-14 | | Release date: | 2015-03-04 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Structural basis for corepressor assembly by the orphan nuclear receptor TLX.
Genes Dev., 29, 2015
|
|
3VIE
 
 | | HIV-gp41 fusion inhibitor Sifuvirtide | | Descriptor: | Envelope glycoprotein gp160, Sifuvirtide | | Authors: | Yao, X, Waltersperger, S, Wang, M.T, Cui, S. | | Deposit date: | 2011-09-29 | | Release date: | 2012-01-18 | | Last modified: | 2024-11-20 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Broad antiviral activity and crystal structure of HIV-1 fusion inhibitor sifuvirtide
J.Biol.Chem., 287, 2012
|
|
3GSM
 
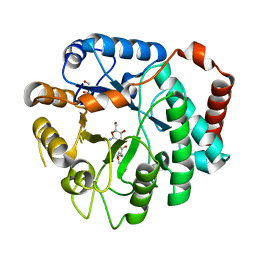 | | Vibrio cholerae family 3 glycoside hydrolase (NagZ) bound to N-Valeryl-PUGNAc | | Descriptor: | Beta-hexosaminidase, GLYCEROL, [[(3R,4R,5S,6R)-4,5-dihydroxy-6-(hydroxymethyl)-3-(pentanoylamino)oxan-2-ylidene]amino] N-phenylcarbamate | | Authors: | Balcewich, M.D, Mark, B.L. | | Deposit date: | 2009-03-27 | | Release date: | 2009-06-30 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Insight into a strategy for attenuating AmpC-mediated beta-lactam resistance: structural basis for selective inhibition of the glycoside hydrolase NagZ.
Protein Sci., 18, 2009
|
|
4XAJ
 
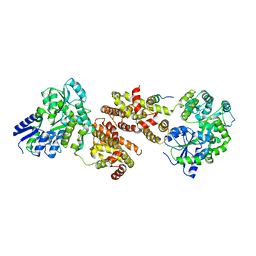 | | Crystal structure of human NR2E1/TLX | | Descriptor: | Atrophin/grunge, Maltose-binding periplasmic protein,Nuclear receptor subfamily 2 group E member 1, alpha-D-glucopyranose-(1-4)-alpha-D-glucopyranose | | Authors: | Zhi, X, Zhou, E, Xu, E. | | Deposit date: | 2014-12-14 | | Release date: | 2015-03-04 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (3.551 Å) | | Cite: | Structural basis for corepressor assembly by the orphan nuclear receptor TLX.
Genes Dev., 29, 2015
|
|
7T4P
 
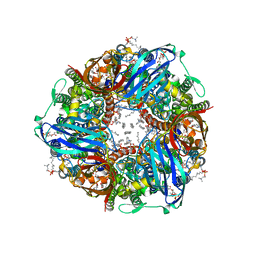 | | CryoEM structure of Methylococcus capsulatus (Bath) pMMO treated with potassium cyanide and copper in a native lipid nanodisc at 3.62 Angstrom resolution | | Descriptor: | 1,2-DIDECANOYL-SN-GLYCERO-3-PHOSPHOCHOLINE, 1,2-dihexanoyl-sn-glycero-3-phosphocholine, Ammonia monooxygenase/methane monooxygenase, ... | | Authors: | Koo, C.W, Rosenzweig, A.C. | | Deposit date: | 2021-12-10 | | Release date: | 2022-03-30 | | Last modified: | 2024-02-28 | | Method: | ELECTRON MICROSCOPY (3.62 Å) | | Cite: | Recovery of particulate methane monooxygenase structure and activity in a lipid bilayer.
Science, 375, 2022
|
|
7T4O
 
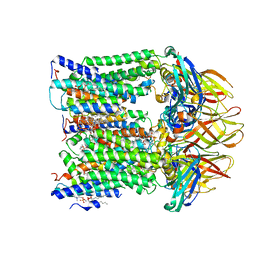 | |
8WRJ
 
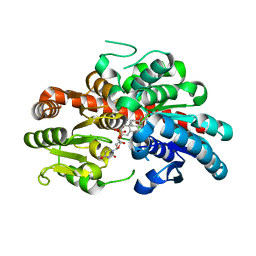 | | glycosyltransferase UGT74AN3 | | Descriptor: | 5-[(1R,2S,4R,6R,7R,10S,11S,14S,16R)-14-hydroxy-7,11-dimethyl-3-oxapentacyclo[8.8.0.02,4.02,7.011,16]octadecan-6-yl]pyran-2-one, Glycosyltransferase, URIDINE-5'-DIPHOSPHATE | | Authors: | Huang, W, Long, F. | | Deposit date: | 2023-10-15 | | Release date: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (1.85 Å) | | Cite: | Substrate Promiscuity, Crystal Structure, and Application of a Plant UDP-Glycosyltransferase UGT74AN3
Acs Catalysis, 14, 2024
|
|
8WRK
 
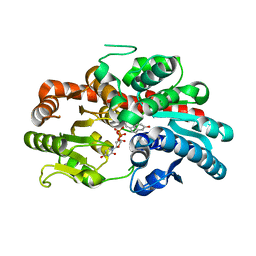 | | glycosyltransferase UGT74AN3 | | Descriptor: | Glycosyltransferase, PICEATANNOL, URIDINE-5'-DIPHOSPHATE | | Authors: | Huang, W, Long, F. | | Deposit date: | 2023-10-15 | | Release date: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (2.03 Å) | | Cite: | Substrate Promiscuity, Crystal Structure, and Application of a Plant UDP-Glycosyltransferase UGT74AN3
Acs Catalysis, 14, 2024
|
|
3GS6
 
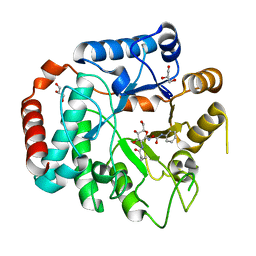 | | Vibrio Cholerea family 3 glycoside hydrolase (NagZ)in complex with N-butyryl-PUGNAc | | Descriptor: | Beta-hexosaminidase, GLYCEROL, [[(3R,4R,5S,6R)-3-(BUTANOYLAMINO)-4,5-DIHYDROXY-6-(HYDROXYMETHYL)OXAN-2-YLIDENE]AMINO] N-PHENYLCARBAMATE | | Authors: | Balcewich, M.D, Mark, B.L. | | Deposit date: | 2009-03-26 | | Release date: | 2009-06-30 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Insight into a strategy for attenuating AmpC-mediated beta-lactam resistance: structural basis for selective inhibition of the glycoside hydrolase NagZ.
Protein Sci., 18, 2009
|
|
7X08
 
 | | S protein of SARS-CoV-2 in complex with 2G1 | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, LINOLEIC ACID, ... | | Authors: | Guo, Y.Y, Zhang, Y.Y, Zhou, Q. | | Deposit date: | 2022-02-21 | | Release date: | 2022-03-09 | | Last modified: | 2024-11-06 | | Method: | ELECTRON MICROSCOPY (2.7 Å) | | Cite: | Broad ultra-potent neutralization of SARS-CoV-2 variants by monoclonal antibodies specific to the tip of RBD.
Cell Discov, 8, 2022
|
|
6NWS
 
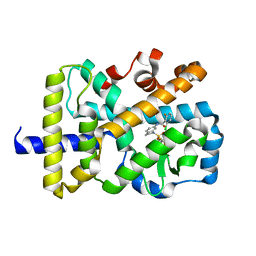 | | RORgamma Ligand Binding Domain | | Descriptor: | 2-chloro-6-fluoro-N-(1-{[3-(trifluoromethyl)phenyl]sulfonyl}-2,3-dihydro-1H-indol-6-yl)benzamide, Nuclear receptor ROR-gamma | | Authors: | Strutzenberg, T.S, Park, H.J, Griffin, P.R. | | Deposit date: | 2019-02-07 | | Release date: | 2019-07-10 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (2.44 Å) | | Cite: | HDX-MS reveals structural determinants for ROR gamma hyperactivation by synthetic agonists.
Elife, 8, 2019
|
|
5CRL
 
 | |
6NWU
 
 | | RORgamma Ligand Binding Domain | | Descriptor: | 6-[(3,5-dichloropyridin-4-yl)methoxy]-1-{[3-(trifluoromethyl)phenyl]sulfonyl}-2,3-dihydro-1H-indole, Nuclear receptor ROR-gamma | | Authors: | Strutzenberg, T.S, Park, H, Griffin, P.R. | | Deposit date: | 2019-02-07 | | Release date: | 2019-07-10 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (3.2 Å) | | Cite: | HDX-MS reveals structural determinants for ROR gamma hyperactivation by synthetic agonists.
Elife, 8, 2019
|
|
6NWT
 
 | | RORgamma Ligand Binding Domain | | Descriptor: | 1,1,1,3,3,3-hexafluoro-2-[2-fluoro-4'-({4-[(pyridin-4-yl)methyl]piperazin-1-yl}methyl)[1,1'-biphenyl]-4-yl]propan-2-ol, Nuclear receptor ROR-gamma | | Authors: | Strutzenberg, T.S, Park, H, Griffin, P.R. | | Deposit date: | 2019-02-07 | | Release date: | 2019-07-10 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (2.35 Å) | | Cite: | HDX-MS reveals structural determinants for ROR gamma hyperactivation by synthetic agonists.
Elife, 8, 2019
|
|
8HJT
 
 | | Crystal Structure of Intracellular B30.2 Domain of VpBTN3 and VpBTN2 in Complex with HMBPP | | Descriptor: | (2E)-4-hydroxy-3-methylbut-2-en-1-yl trihydrogen diphosphate, 4-(2-HYDROXYETHYL)-1-PIPERAZINE ETHANESULFONIC ACID, Butyrophylin 3, ... | | Authors: | Yang, Y.Y, Shen, P.P, Li, X, Yi, S.M, Zhang, M.T, Huang, J.-W, Chen, C.-C, Guo, R.-T. | | Deposit date: | 2022-11-23 | | Release date: | 2023-09-13 | | Last modified: | 2024-11-27 | | Method: | X-RAY DIFFRACTION (2.91 Å) | | Cite: | Phosphoantigens glue butyrophilin 3A1 and 2A1 to activate V gamma 9V delta 2 T cells.
Nature, 621, 2023
|
|
5BWA
 
 | | Crystal structure of ODC-PLP-AZ1 ternary complex | | Descriptor: | Ornithine decarboxylase, Ornithine decarboxylase antizyme 1, PYRIDOXAL-5'-PHOSPHATE | | Authors: | Wu, D.H. | | Deposit date: | 2015-06-07 | | Release date: | 2015-12-09 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (3.2 Å) | | Cite: | Structural basis of Ornithine Decarboxylase inactivation and accelerated degradation by polyamine sensor Antizyme1
Sci Rep, 5, 2015
|
|
8RVF
 
 | | CRYSTAL STRUCTURE OF HUMAN MONOGLYCERIDE LIPASE IN COMPLEX WITH COMPOUND 5 | | Descriptor: | 1,2-ETHANEDIOL, Monoglyceride lipase, [6-[[2,2-bis(fluoranyl)-1$l^{4},3-diaza-2$l^{4}-boratricyclo[7.3.0.0^{3,7}]dodeca-1(12),4,6,8,10-pentaen-8-yl]methyl]-2-azaspiro[3.3]heptan-2-yl]-[6-(3-cyclopropyl-1,2,4-triazol-1-yl)-2-azaspiro[3.3]heptan-2-yl]methanone | | Authors: | Leibrock, L, Hentsch, A, Nazare, M, Grether, U, Kuhn, B, Blaising, J, Benz, J. | | Deposit date: | 2024-02-01 | | Release date: | 2025-02-12 | | Last modified: | 2025-04-09 | | Method: | X-RAY DIFFRACTION (1.43 Å) | | Cite: | Highly Specific Miniaturized Fluorescent Monoacylglycerol Lipase Probes Enable Translational Research.
J.Am.Chem.Soc., 147, 2025
|
|
8YZD
 
 | | Structure of JN.1 RBD protein in complex with ACE2. | | Descriptor: | Angiotensin-converting enzyme 2, Spike glycoprotein,Fibritin,fusion protein | | Authors: | Wang, Y.J, Zhang, X, Sun, L. | | Deposit date: | 2024-04-06 | | Release date: | 2025-01-22 | | Last modified: | 2025-07-23 | | Method: | ELECTRON MICROSCOPY (3.07 Å) | | Cite: | Lineage-specific pathogenicity, immune evasion, and virological features of SARS-CoV-2 BA.2.86/JN.1 and EG.5.1/HK.3.
Nat Commun, 15, 2024
|
|
