5X0S
 
 | | Solution NMR structure of peptide toxin SsTx from Scolopendra subspinipes mutilans | | Descriptor: | SsTx | | Authors: | Wu, F, Luo, L, Qu, D, Zhang, L, Tian, C, Lai, R. | | Deposit date: | 2017-01-23 | | Release date: | 2018-01-24 | | Last modified: | 2024-10-23 | | Method: | SOLUTION NMR | | Cite: | Centipedes subdue giant prey by blocking KCNQ channels
Proc. Natl. Acad. Sci. U.S.A., 115, 2018
|
|
7E1Q
 
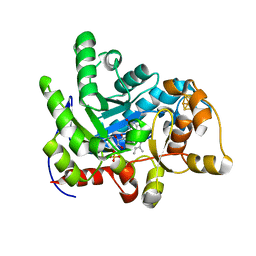 | | Crystal structure of dehydrogenase/isomerase FabX from Helicobacter pylori | | Descriptor: | 2-nitropropane dioxygenase, CHLORIDE ION, FLAVIN MONONUCLEOTIDE, ... | | Authors: | Zhou, J.S, Zhang, L, Zhang, L. | | Deposit date: | 2021-02-03 | | Release date: | 2021-12-01 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Helicobacter pylori FabX contains a [4Fe-4S] cluster essential for unsaturated fatty acid synthesis.
Nat Commun, 12, 2021
|
|
7E1R
 
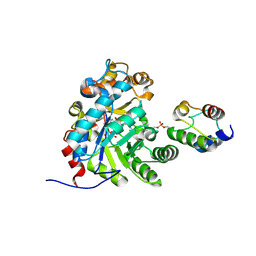 | | Crystal structure of Dehydrogenase/isomerase FabX from Helicobacter pylori in complex with holo-ACP | | Descriptor: | 2-nitropropane dioxygenase, Acyl carrier protein,Acyl carrier protein, FLAVIN MONONUCLEOTIDE, ... | | Authors: | Zhou, J.S, Zhang, L, Zhang, L. | | Deposit date: | 2021-02-03 | | Release date: | 2021-12-01 | | Last modified: | 2024-11-20 | | Method: | X-RAY DIFFRACTION (2.795 Å) | | Cite: | Helicobacter pylori FabX contains a [4Fe-4S] cluster essential for unsaturated fatty acid synthesis.
Nat Commun, 12, 2021
|
|
7E1S
 
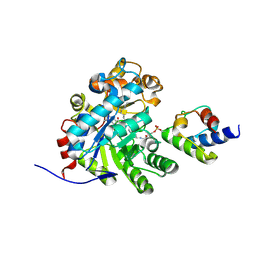 | | Crystal structure of dehydrogenase/isomerase FabX from Helicobacter pylori in complex with octanoyl-ACP | | Descriptor: | 2-nitropropane dioxygenase, Acyl carrier protein,Acyl carrier protein, FLAVIN MONONUCLEOTIDE, ... | | Authors: | Zhou, J.S, Zhang, L, Zhang, L. | | Deposit date: | 2021-02-03 | | Release date: | 2021-12-01 | | Last modified: | 2024-11-13 | | Method: | X-RAY DIFFRACTION (2.31 Å) | | Cite: | Helicobacter pylori FabX contains a [4Fe-4S] cluster essential for unsaturated fatty acid synthesis.
Nat Commun, 12, 2021
|
|
2M0Q
 
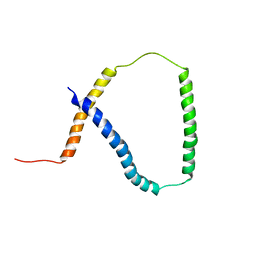 | | Solution NMR analysis of intact KCNE2 in detergent micelles demonstrate a straight transmembrane helix | | Descriptor: | Potassium voltage-gated channel subfamily E member 2 | | Authors: | Lai, C, Li, P, Chen, L, Zhang, L, Wu, F, Tian, C. | | Deposit date: | 2012-11-01 | | Release date: | 2014-04-30 | | Last modified: | 2024-05-15 | | Method: | SOLUTION NMR | | Cite: | Differential modulations of KCNQ1 by auxiliary proteins KCNE1 and KCNE2.
Sci Rep, 4, 2014
|
|
3VPN
 
 | | Crystal structure of human ribonucleotide reductase subunit M2 (hRRM2) mutant | | Descriptor: | FE (III) ION, MAGNESIUM ION, Ribonucleoside-diphosphate reductase subunit M2 | | Authors: | Chen, X, Xu, Z, Liu, H, Zhang, L, Chen, B, Zhu, L, Yang, C, Zhu, W, Shao, J. | | Deposit date: | 2012-03-05 | | Release date: | 2013-03-06 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (2.25 Å) | | Cite: | Essential role of E106 in the proton-coupled electron transfer in human
to be published
|
|
6KX9
 
 | |
6IHC
 
 | | Crystal structure of (3R)-Hydroxyacyl-Acyl Carrier Protein Dehydratase(FabZ) Y100A mutant in complex with holo-ACP from Helicobacter pylori | | Descriptor: | 3-hydroxyacyl-[acyl-carrier-protein] dehydratase FabZ, CITRIC ACID, N~3~-[(2S)-2-hydroxy-3,3-dimethyl-4-(phosphonooxy)butanoyl]-N-(2-sulfanylethyl)-beta-alaninamide, ... | | Authors: | Shen, S.Q, Zhang, L, Zhang, L. | | Deposit date: | 2018-09-29 | | Release date: | 2019-04-10 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | A back-door Phenylalanine coordinates the stepwise hexameric loading of acyl carrier protein by the fatty acid biosynthesis enzyme beta-hydroxyacyl-acyl carrier protein dehydratase (FabZ).
Int. J. Biol. Macromol., 128, 2019
|
|
8K1O
 
 | | mycobacterial efflux pump, AMPPNP bound state | | Descriptor: | CARDIOLIPIN, MAGNESIUM ION, Multidrug efflux system ATP-binding protein Rv1218c, ... | | Authors: | Wang, Y, Wu, F, Zhang, L, Rao, Z. | | Deposit date: | 2023-07-11 | | Release date: | 2024-07-17 | | Last modified: | 2025-01-29 | | Method: | ELECTRON MICROSCOPY (2.9 Å) | | Cite: | Cryo-EM structures of a mycobacterial ABC transporter that mediates rifampicin resistance.
Proc.Natl.Acad.Sci.USA, 121, 2024
|
|
8K1M
 
 | | mycobacterial efflux pump, apo state | | Descriptor: | (1S)-2-{[(S)-(2-aminoethoxy)(hydroxy)phosphoryl]oxy}-1-[(octadecanoyloxy)methyl]ethyl (9Z)-octadec-9-enoate, CARDIOLIPIN, Multidrug efflux system ATP-binding protein Rv1218c, ... | | Authors: | Wang, Y, Wu, F, Zhang, L, Rao, Z. | | Deposit date: | 2023-07-11 | | Release date: | 2024-07-17 | | Last modified: | 2025-01-29 | | Method: | ELECTRON MICROSCOPY (2.9 Å) | | Cite: | Cryo-EM structures of a mycobacterial ABC transporter that mediates rifampicin resistance.
Proc.Natl.Acad.Sci.USA, 121, 2024
|
|
8K1N
 
 | | mycobacterial efflux pump, substrate-bound state | | Descriptor: | CARDIOLIPIN, Multidrug efflux system ATP-binding protein Rv1218c, Multidrug efflux system permease protein Rv1217c, ... | | Authors: | Wang, Y, Wu, F, Zhang, L, Rao, Z. | | Deposit date: | 2023-07-11 | | Release date: | 2024-07-17 | | Last modified: | 2025-01-29 | | Method: | ELECTRON MICROSCOPY (3 Å) | | Cite: | Cryo-EM structures of a mycobacterial ABC transporter that mediates rifampicin resistance.
Proc.Natl.Acad.Sci.USA, 121, 2024
|
|
7CFN
 
 | | Cryo-EM structure of the INT-777-bound GPBAR-Gs complex | | Descriptor: | (2S,4R)-4-[(3R,5S,6R,7R,8R,9S,10S,12S,13R,14S,17R)-6-ethyl-10,13-dimethyl-3,7,12-tris(oxidanyl)-2,3,4,5,6,7,8,9,11,12,14,15,16,17-tetradecahydro-1H-cyclopenta[a]phenanthren-17-yl]-2-methyl-pentanoic acid, CHOLESTEROL, G-protein coupled bile acid receptor 1, ... | | Authors: | Yang, F, Mao, C, Guo, L, Lin, J, Ming, Q, Xiao, P, Wu, X, Shen, Q, Guo, S, Shen, D, Lu, R, Zhang, L, Huang, S, Ping, Y, Zhang, C, Ma, C, Zhang, K, Liang, X, Shen, Y, Nan, F, Yi, F, Luca, V, Zhou, J, Jiang, C, Sun, J, Xie, X, Yu, X, Zhang, Y. | | Deposit date: | 2020-06-27 | | Release date: | 2020-09-09 | | Last modified: | 2024-11-20 | | Method: | ELECTRON MICROSCOPY (3 Å) | | Cite: | Structural basis of GPBAR activation and bile acid recognition.
Nature, 587, 2020
|
|
7C80
 
 | | E30 F-particle in complex with 4B10 | | Descriptor: | Heavy chain, Light chain, SPHINGOSINE, ... | | Authors: | Wang, K, Zheng, B, Zhang, L, Cui, L, Su, X, Zhang, Q, Guo, Y, Zhu, L, Zhu, F, Rao, Z, Wang, X. | | Deposit date: | 2020-05-28 | | Release date: | 2020-07-29 | | Last modified: | 2025-04-09 | | Method: | ELECTRON MICROSCOPY (3.7 Å) | | Cite: | Serotype specific epitopes identified by neutralizing antibodies underpin immunogenic differences in Enterovirus B.
Nat Commun, 11, 2020
|
|
7C81
 
 | | E30 F-particle in complex with 6C5 | | Descriptor: | Heavy chain, Light chain, SPHINGOSINE, ... | | Authors: | Wang, K, Zheng, B, Zhang, L, Cui, L, Su, X, Zhang, Q, Guo, Y, Zhu, L, Zhu, F, Rao, Z, Wang, X. | | Deposit date: | 2020-05-28 | | Release date: | 2020-07-29 | | Last modified: | 2025-04-09 | | Method: | ELECTRON MICROSCOPY (3.1 Å) | | Cite: | Serotype specific epitopes identified by neutralizing antibodies underpin immunogenic differences in Enterovirus B.
Nat Commun, 11, 2020
|
|
7CDI
 
 | | Crystal structure of SARS-CoV-2 antibody P2C-1F11 with RBD | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, Spike protein S1, antibody P2C-1F11 heavy chain, ... | | Authors: | Wang, X, Zhang, L, Ge, J, Wang, R. | | Deposit date: | 2020-06-19 | | Release date: | 2020-11-18 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (2.96 Å) | | Cite: | Antibody neutralization of SARS-CoV-2 through ACE2 receptor mimicry.
Nat Commun, 12, 2021
|
|
7CDJ
 
 | | Crystal structure of SARS-CoV-2 antibody P2C-1A3 with RBD | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, Spike protein S1, antibody P2C-1A3 heavy chain, ... | | Authors: | Wang, X, Zhang, L, Ge, J, Wang, R. | | Deposit date: | 2020-06-19 | | Release date: | 2020-11-18 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (3.396 Å) | | Cite: | Antibody neutralization of SARS-CoV-2 through ACE2 receptor mimicry.
Nat Commun, 12, 2021
|
|
7CFM
 
 | | Cryo-EM structure of the P395-bound GPBAR-Gs complex | | Descriptor: | 2-(ethylamino)-6-[3-(4-propan-2-ylphenyl)propanoyl]-7,8-dihydro-5H-pyrido[4,3-d]pyrimidine-4-carboxamide, CHOLESTEROL, G-protein coupled bile acid receptor 1, ... | | Authors: | Yang, F, Mao, C, Guo, L, Lin, J, Ming, Q, Xiao, P, Wu, X, Shen, Q, Guo, S, Shen, D, Lu, R, Zhang, L, Huang, S, Ping, Y, Zhang, C, Ma, C, Zhang, K, Liang, X, Shen, Y, Nan, F, Yi, F, Luca, V, Zhou, J, Jiang, C, Sun, J, Xie, X, Yu, X, Zhang, Y. | | Deposit date: | 2020-06-27 | | Release date: | 2020-09-09 | | Last modified: | 2024-10-09 | | Method: | ELECTRON MICROSCOPY (3 Å) | | Cite: | Structural basis of GPBAR activation and bile acid recognition.
Nature, 587, 2020
|
|
2K8P
 
 | | Characterisation of the structural features and interactions of sclerostin: molecular insight into a key regulator of Wnt-mediated bone formation | | Descriptor: | Sclerostin | | Authors: | Veverka, V, Henry, A.J, Slocombe, P.M, Ventom, A, Mulloy, B, Muskett, F.W, Muzylak, M, Greenslade, K, Moore, A, Zhang, L, Gong, J, Qian, X, Paszty, C, Taylor, R.J, Robinson, M.K, Carr, M.D. | | Deposit date: | 2008-09-18 | | Release date: | 2009-02-17 | | Last modified: | 2024-11-06 | | Method: | SOLUTION NMR | | Cite: | Characterization of the Structural Features and Interactions of Sclerostin: Molecular insight into a key regulator of Wnt-mediated bone formation
J.Biol.Chem., 284, 2009
|
|
8YFU
 
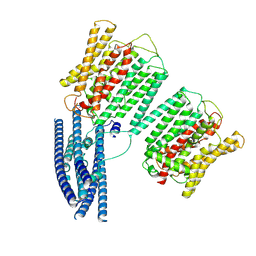 | | Cryo EM structure of human phosphate channel XPR1 at intermediate state | | Descriptor: | Solute carrier family 53 member 1 | | Authors: | Lu, Y, Yue, C, Zhang, L, Yao, D, Yu, Y, Cao, Y. | | Deposit date: | 2024-02-25 | | Release date: | 2024-10-09 | | Last modified: | 2024-12-18 | | Method: | ELECTRON MICROSCOPY (4.59 Å) | | Cite: | Structural basis for inositol pyrophosphate gating of the phosphate channel XPR1.
Science, 386, 2024
|
|
8YET
 
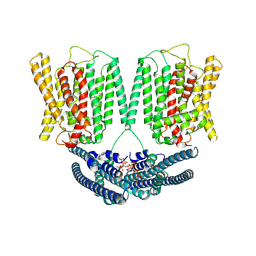 | | Cryo EM structure of human phosphate channel XPR1 in complex with IP6 | | Descriptor: | INOSITOL HEXAKISPHOSPHATE, Solute carrier family 53 member 1 | | Authors: | Lu, Y, Yue, C, Zhang, L, Yao, D, Yu, Y, Cao, Y. | | Deposit date: | 2024-02-23 | | Release date: | 2024-10-09 | | Last modified: | 2024-12-18 | | Method: | ELECTRON MICROSCOPY (4.16 Å) | | Cite: | Structural basis for inositol pyrophosphate gating of the phosphate channel XPR1.
Science, 386, 2024
|
|
8YFX
 
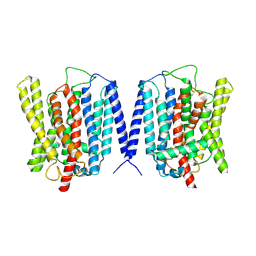 | | Cryo EM structure of human phosphate channel XPR1 at inward-facing state | | Descriptor: | Solute carrier family 53 member 1 | | Authors: | Lu, Y, Yue, C, Zhang, L, Yao, D, Yu, Y, Cao, Y. | | Deposit date: | 2024-02-25 | | Release date: | 2024-10-09 | | Last modified: | 2024-12-18 | | Method: | ELECTRON MICROSCOPY (3.56 Å) | | Cite: | Structural basis for inositol pyrophosphate gating of the phosphate channel XPR1.
Science, 386, 2024
|
|
8YFW
 
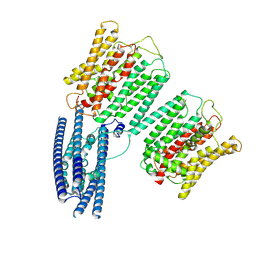 | | Cryo EM structure of human phosphate channel XPR1 at intermediate state | | Descriptor: | Solute carrier family 53 member 1 | | Authors: | Lu, Y, Yue, C, Zhang, L, Yao, D, Yu, Y, Cao, Y. | | Deposit date: | 2024-02-25 | | Release date: | 2024-10-09 | | Last modified: | 2024-12-18 | | Method: | ELECTRON MICROSCOPY (3.65 Å) | | Cite: | Structural basis for inositol pyrophosphate gating of the phosphate channel XPR1.
Science, 386, 2024
|
|
8YEX
 
 | | Cryo EM structure of human phosphate channel XPR1 at apo state | | Descriptor: | Solute carrier family 53 member 1 | | Authors: | Lu, Y, Yue, C, Zhang, L, Yao, D, Yu, Y, Cao, Y. | | Deposit date: | 2024-02-23 | | Release date: | 2024-10-09 | | Last modified: | 2024-12-18 | | Method: | ELECTRON MICROSCOPY (3.68 Å) | | Cite: | Structural basis for inositol pyrophosphate gating of the phosphate channel XPR1.
Science, 386, 2024
|
|
8YF4
 
 | | Cryo EM structure of human phosphate channel XPR1 at open and inward-facing state | | Descriptor: | CHOLESTEROL HEMISUCCINATE, Solute carrier family 53 member 1 | | Authors: | Lu, Y, Yue, C, Zhang, L, Yao, D, Yu, Y, Cao, Y. | | Deposit date: | 2024-02-24 | | Release date: | 2024-10-09 | | Last modified: | 2024-12-18 | | Method: | ELECTRON MICROSCOPY (3.41 Å) | | Cite: | Structural basis for inositol pyrophosphate gating of the phosphate channel XPR1.
Science, 386, 2024
|
|
8YFD
 
 | | Cryo EM structure of human phosphate channel XPR1 at open state | | Descriptor: | Solute carrier family 53 member 1 | | Authors: | Lu, Y, Yue, C, Zhang, L, Yao, D, Yu, Y, Cao, Y. | | Deposit date: | 2024-02-24 | | Release date: | 2024-10-09 | | Last modified: | 2024-12-18 | | Method: | ELECTRON MICROSCOPY (3.57 Å) | | Cite: | Structural basis for inositol pyrophosphate gating of the phosphate channel XPR1.
Science, 386, 2024
|
|
