6UEY
 
 | | Pistol ribozyme transition-state analog vanadate | | Descriptor: | MAGNESIUM ION, RNA (5'-R(*UP*CP*UP*GP*CP*UP*CP*UP*CP*(GVA)*UP*CP*CP*AP*A)-3'), RNA (50-MER) | | Authors: | Teplova, M, Falschlunger, C, Krasheninina, O, Patel, D.J, Micura, R. | | Deposit date: | 2019-09-23 | | Release date: | 2019-12-18 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Crucial Roles of Two Hydrated Mg2+Ions in Reaction Catalysis of the Pistol Ribozyme.
Angew.Chem.Int.Ed.Engl., 59, 2020
|
|
6V9Q
 
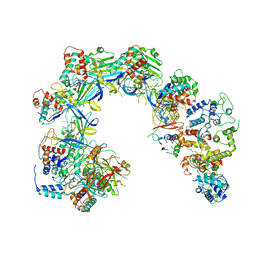 | | Cryo-EM structure of Cascade-TniQ binary complex | | Descriptor: | Cas8, RNA (61-MER), TniQ family protein, ... | | Authors: | Jia, N, Patel, D.J. | | Deposit date: | 2019-12-15 | | Release date: | 2020-01-29 | | Last modified: | 2024-03-06 | | Method: | ELECTRON MICROSCOPY (2.9 Å) | | Cite: | Structure-function insights into the initial step of DNA integration by a CRISPR-Cas-Transposon complex.
Cell Res., 30, 2020
|
|
6VBW
 
 | |
6VRC
 
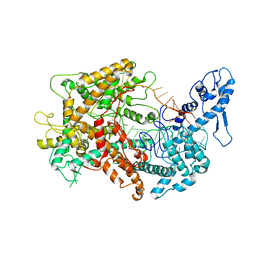 | | Cryo-EM structure of Cas13(crRNA) | | Descriptor: | CRISPR-associated endoribonuclease Cas13a, RNA (51-MER) | | Authors: | Jia, N, Meeske, A.J, Marraffini, L.A, Patel, D.J. | | Deposit date: | 2020-02-07 | | Release date: | 2020-06-10 | | Last modified: | 2024-03-06 | | Method: | ELECTRON MICROSCOPY (3.2 Å) | | Cite: | A phage-encoded anti-CRISPR enables complete evasion of type VI-A CRISPR-Cas immunity.
Science, 369, 2020
|
|
6UFK
 
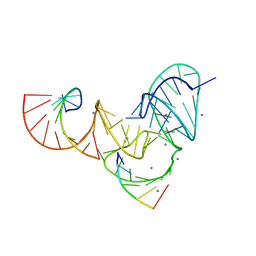 | | Pistol ribozyme product crystal soaked in Mn2+ | | Descriptor: | MANGANESE (II) ION, RNA (5'-R(*UP*CP*CP*AP*G)-3'), RNA (5'-R(*UP*CP*UP*GP*CP*UP*CP*UP*CP*(23G))-3'), ... | | Authors: | Teplova, M, Falschlunger, C, Krasheninina, O, Patel, D.J, Micura, R. | | Deposit date: | 2019-09-24 | | Release date: | 2019-12-18 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (3.2 Å) | | Cite: | Crucial Roles of Two Hydrated Mg2+Ions in Reaction Catalysis of the Pistol Ribozyme.
Angew.Chem.Int.Ed.Engl., 59, 2020
|
|
6V9P
 
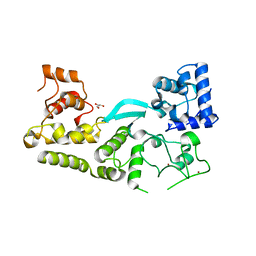 | | Crystal structure of the transposition protein TniQ | | Descriptor: | GLYCEROL, TniQ family protein, ZINC ION | | Authors: | Jia, N, Patel, D.J. | | Deposit date: | 2019-12-14 | | Release date: | 2020-01-29 | | Last modified: | 2024-03-06 | | Method: | X-RAY DIFFRACTION (1.93 Å) | | Cite: | Structure-function insights into the initial step of DNA integration by a CRISPR-Cas-Transposon complex.
Cell Res., 30, 2020
|
|
6VRB
 
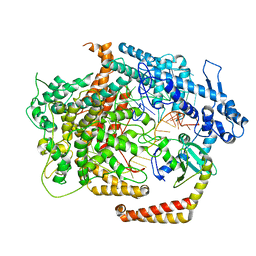 | | Cryo-EM structure of AcrVIA1-Cas13(crRNA) complex | | Descriptor: | AcrVIA1, CRISPR-associated endoribonuclease Cas13a, RNA (52-MER) | | Authors: | Jia, N, Meeske, A.J, Marraffini, L.A, Patel, D.J. | | Deposit date: | 2020-02-07 | | Release date: | 2020-06-10 | | Last modified: | 2024-03-06 | | Method: | ELECTRON MICROSCOPY (3 Å) | | Cite: | A phage-encoded anti-CRISPR enables complete evasion of type VI-A CRISPR-Cas immunity.
Science, 369, 2020
|
|
2OE5
 
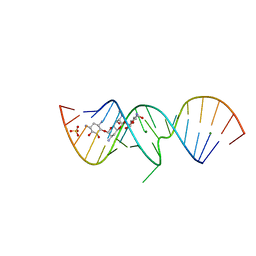 | | 1.5 A X-ray crystal structure of Apramycin complex with RNA fragment GGCGUCGCUAGUACCG/GGUACUAAAAGUCGCCC containing the human ribosomal decoding A site: RNA construct with 3'-overhang | | Descriptor: | APRAMYCIN, MAGNESIUM ION, RNA (5'-R(*GP*GP*CP*GP*UP*CP*GP*CP*UP*AP*GP*UP*AP*CP*CP*G)-3'), ... | | Authors: | Hermann, T, Tereshko, V, Skripkin, E, Patel, D.J. | | Deposit date: | 2006-12-28 | | Release date: | 2007-02-13 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (1.51 Å) | | Cite: | Apramycin recognition by the human ribosomal decoding site.
Blood Cells Mol.Dis., 38, 2007
|
|
2O3M
 
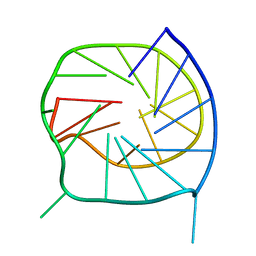 | | Monomeric G-DNA tetraplex from human C-kit promoter | | Descriptor: | 5'-D(*AP*GP*GP*GP*AP*GP*GP*GP*CP*GP*CP*TP*GP*GP*GP*AP*GP*GP*AP*GP*GP*G)-3' | | Authors: | Phan, A.T, Kuryavyi, V.V, Burge, S, Neidle, S, Patel, D.J. | | Deposit date: | 2006-12-01 | | Release date: | 2007-05-22 | | Last modified: | 2023-12-27 | | Method: | SOLUTION NMR | | Cite: | Structure of an unprecedented G-quadruplex scaffold in the human c-kit promoter.
J.Am.Chem.Soc., 129, 2007
|
|
2OE8
 
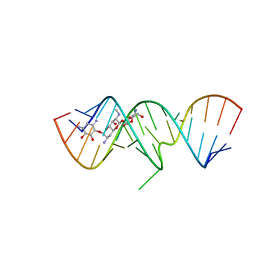 | | 1.8 A X-ray crystal structure of Apramycin complex with RNA fragment GGGCGUCGCUAGUACC/CGGUACUAAAAGUCGCC containing the human ribosomal decoding A site: RNA construct with 5'-overhang | | Descriptor: | APRAMYCIN, RNA (5'-R(*CP*GP*GP*UP*AP*CP*UP*AP*AP*AP*AP*GP*UP*CP*GP*CP*C)-3'), RNA (5'-R(*GP*GP*GP*CP*GP*UP*CP*GP*CP*UP*AP*GP*UP*AP*CP*C)-3') | | Authors: | Hermann, T, Tereshko, V, Skripkin, E, Patel, D.J. | | Deposit date: | 2006-12-28 | | Release date: | 2007-02-13 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Apramycin recognition by the human ribosomal decoding site.
Blood Cells Mol.Dis., 38, 2007
|
|
2TOB
 
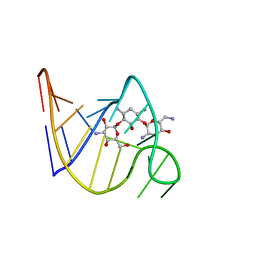 | | SOLUTION STRUCTURE OF THE TOBRAMYCIN-RNA APTAMER COMPLEX, NMR, 13 STRUCTURES | | Descriptor: | 1,3-DIAMINO-4,5,6-TRIHYDROXY-CYCLOHEXANE, 2,6-diammonio-2,3,6-trideoxy-alpha-D-glucopyranose, 3-ammonio-3-deoxy-alpha-D-glucopyranose, ... | | Authors: | Jiang, L, Patel, D.J. | | Deposit date: | 1998-06-17 | | Release date: | 1999-06-22 | | Last modified: | 2024-05-01 | | Method: | SOLUTION NMR | | Cite: | Solution structure of the tobramycin-RNA aptamer complex.
Nat.Struct.Biol., 5, 1998
|
|
2RI5
 
 | |
2RHU
 
 | |
2RI7
 
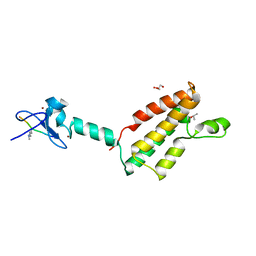 | |
2RI2
 
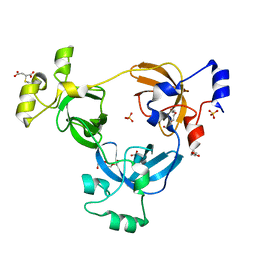 | |
2RHI
 
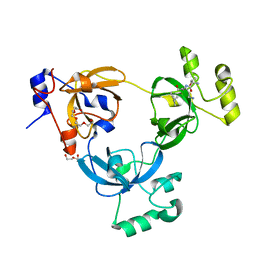 | |
2RHY
 
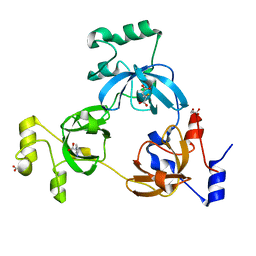 | |
2RHX
 
 | |
2RI3
 
 | |
2RHZ
 
 | |
4QLN
 
 | | structure of ydao riboswitch binding with c-di-dAMP | | Descriptor: | (2R,3R,3aS,5R,7aR,9R,10R,10aS,12R,14aR)-2,9-bis(6-amino-9H-purin-9-yl)octahydro-2H,7H-difuro[3,2-d:3',2'-j][1,3,7,9,2,8 ]tetraoxadiphosphacyclododecine-3,5,10,12-tetrol 5,12-dioxide, MAGNESIUM ION, RNA (117-MER) | | Authors: | Ren, A.M, Patel, D.J. | | Deposit date: | 2014-06-12 | | Release date: | 2014-08-13 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (2.65 Å) | | Cite: | c-di-AMP binds the ydaO riboswitch in two pseudo-symmetry-related pockets.
Nat.Chem.Biol., 10, 2014
|
|
4QXH
 
 | |
4QXB
 
 | |
4QXR
 
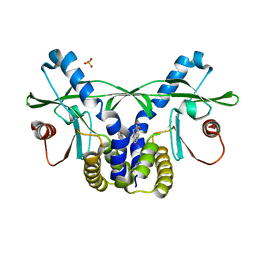 | | Crystal structure of hSTING(S162A/G230I/Q266I) in complex with DMXAA | | Descriptor: | (5,6-dimethyl-9-oxo-9H-xanthen-4-yl)acetic acid, SULFATE ION, Stimulator of interferon genes protein | | Authors: | Gao, P, Patel, D.J. | | Deposit date: | 2014-07-21 | | Release date: | 2014-09-10 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (2.37 Å) | | Cite: | Binding-Pocket and Lid-Region Substitutions Render Human STING Sensitive to the Species-Specific Drug DMXAA.
Cell Rep, 8, 2014
|
|
4QEO
 
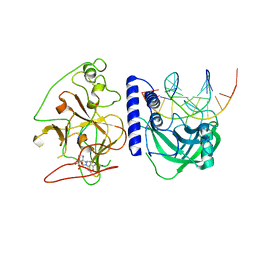 | | crystal structure of KRYPTONITE in complex with mCHH DNA, H3(1-15) peptide and SAH | | Descriptor: | DNA 5'-ACTGATGAGTACCAT-3', DNA 5'-GGTACT(5CM)ATCAGTAT-3', Histone H3, ... | | Authors: | Du, J, Li, S, Patel, D.J. | | Deposit date: | 2014-05-17 | | Release date: | 2014-07-30 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Mechanism of DNA Methylation-Directed Histone Methylation by KRYPTONITE.
Mol.Cell, 55, 2014
|
|
