4YDK
 
 | | Crystal structure of broadly and potently neutralizing antibody C38-VRC16.01 in complex with HIV-1 clade AE strain 93TH057 gp120 | | Descriptor: | 2-[N-CYCLOHEXYLAMINO]ETHANE SULFONIC ACID, 2-acetamido-2-deoxy-beta-D-glucopyranose, DI(HYDROXYETHYL)ETHER, ... | | Authors: | Zhou, T, Moquin, S, Zheng, A, Kwong, P.D. | | Deposit date: | 2015-02-22 | | Release date: | 2015-06-03 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (2.051 Å) | | Cite: | Structural Repertoire of HIV-1-Neutralizing Antibodies Targeting the CD4 Supersite in 14 Donors.
Cell, 161, 2015
|
|
4YDJ
 
 | | Crystal structure of broadly and potently neutralizing antibody 44-VRC13.01 in complex with HIV-1 clade AE strain 93TH057 gp120 | | Descriptor: | (R,R)-2,3-BUTANEDIOL, 2-acetamido-2-deoxy-beta-D-glucopyranose, CHLORIDE ION, ... | | Authors: | Zhou, T, Moquin, S, Kwong, P.D. | | Deposit date: | 2015-02-22 | | Release date: | 2015-06-03 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (2.308 Å) | | Cite: | Structural Repertoire of HIV-1-Neutralizing Antibodies Targeting the CD4 Supersite in 14 Donors.
Cell, 161, 2015
|
|
4YDL
 
 | | Crystal structure of broadly and potently neutralizing antibody C38-VRC18.02 in complex with HIV-1 clade AE strain 93TH057gp120 | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 4-(2-HYDROXYETHYL)-1-PIPERAZINE ETHANESULFONIC ACID, Envelope glycoprotein gp160,Envelope glycoprotein gp160, ... | | Authors: | Zhou, T, Srivatsan, S, Kwong, P.D. | | Deposit date: | 2015-02-22 | | Release date: | 2015-06-03 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Structural Repertoire of HIV-1-Neutralizing Antibodies Targeting the CD4 Supersite in 14 Donors.
Cell, 161, 2015
|
|
4YE4
 
 | | Crystal Structure of Neutralizing Antibody HJ16 in Complex with HIV-1 gp120 | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, HT593.1 gp120, Heavy chain human antibody HJ16, ... | | Authors: | Kwong, P.D, Chen, L, Zhou, T. | | Deposit date: | 2015-02-23 | | Release date: | 2015-07-22 | | Last modified: | 2022-03-30 | | Method: | X-RAY DIFFRACTION (2.72 Å) | | Cite: | Structural Repertoire of HIV-1-Neutralizing Antibodies Targeting the CD4 Supersite in 14 Donors.
Cell, 161, 2015
|
|
7E2P
 
 | | The Crystal Structure of Mycoplasma bovis enolase | | Descriptor: | Enolase | | Authors: | Chen, R, Zhang, S, Gan, R, Wang, W, Ran, T, Shao, G, Xiong, Q, Feng, Z. | | Deposit date: | 2021-02-07 | | Release date: | 2022-02-02 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Evidence for the Rapid and Divergent Evolution of Mycoplasmas: Structural and Phylogenetic Analysis of Enolases.
Front Mol Biosci, 8, 2022
|
|
7E2Q
 
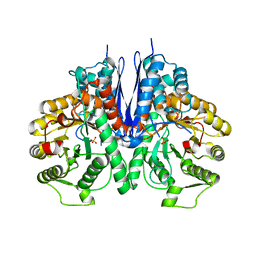 | | Crystal structure of Mycoplasma pneumoniae Enolase | | Descriptor: | Enolase, SULFATE ION | | Authors: | Chen, R, Zhang, S, Gan, R, Wang, W, Ran, T, Xiong, Q, Shao, G, Feng, Z. | | Deposit date: | 2021-02-07 | | Release date: | 2022-02-02 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Evidence for the Rapid and Divergent Evolution of Mycoplasmas: Structural and Phylogenetic Analysis of Enolases.
Front Mol Biosci, 8, 2022
|
|
4YFL
 
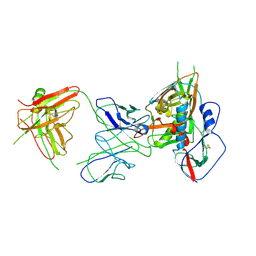 | | Crystal structure of VH1-46 germline-derived CD4-binding site-directed antibody 1B2530 in complex with HIV-1 clade A/E 93TH057 gp120 | | Descriptor: | 1B2530 Light chain, 1B2530 heavy chain, 2-acetamido-2-deoxy-beta-D-glucopyranose, ... | | Authors: | Acharya, P, Zhou, T, Moquin, S, Kwong, P.D. | | Deposit date: | 2015-02-25 | | Release date: | 2015-06-03 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (3.387 Å) | | Cite: | Structural Repertoire of HIV-1-Neutralizing Antibodies Targeting the CD4 Supersite in 14 Donors.
Cell, 161, 2015
|
|
4A5W
 
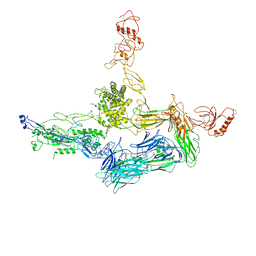 | | Crystal structure of C5b6 | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, CALCIUM ION, COMPLEMENT C5, ... | | Authors: | Hadders, M.A, Bubeck, D, Forneris, F, Pangburn, M, Llorca, O, Lea, S.M, Gros, P. | | Deposit date: | 2011-10-28 | | Release date: | 2012-03-14 | | Last modified: | 2024-07-24 | | Method: | X-RAY DIFFRACTION (3.5 Å) | | Cite: | Assembly and Regulation of the Membrane Attack Complex Based on Structures of C5B6 and Sc5B9.
Cell Rep., 1, 2012
|
|
4BQR
 
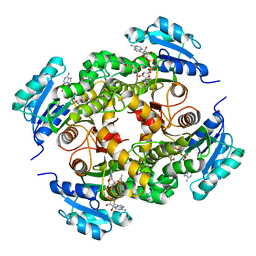 | | Mtb InhA complex with Methyl-thiazole compound 11 | | Descriptor: | (NZ)-2-[2,6-bis(fluoranyl)phenyl]-N-[5-[(1S)-1-(4-methyl-1,3-thiazol-2-yl)-1-oxidanyl-ethyl]-3H-1,3,4-thiadiazol-2-ylidene]ethanamide, ENOYL-[ACYL-CARRIER-PROTEIN] REDUCTASE [NADH], NICOTINAMIDE-ADENINE-DINUCLEOTIDE | | Authors: | Read, J.A, Gingell, H, Madhavapeddi, P, Shirude, P.S. | | Deposit date: | 2013-05-31 | | Release date: | 2013-12-11 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (2.05 Å) | | Cite: | Methyl-Thiazoles: A Novel Mode of Inhibition with the Potential to Develop Novel Inhibitors Targeting Inha in Mycobacterium Tuberculosis.
J.Med.Chem., 56, 2013
|
|
4BQP
 
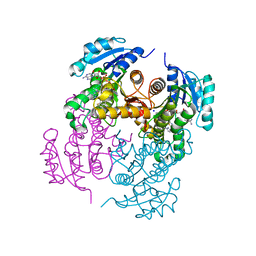 | | Mtb InhA complex with Methyl-thiazole compound 7 | | Descriptor: | (1S)-1-(5-{[1-(2,6-DIFLUOROBENZYL)-1H-PYRAZOL-3-YL]AMINO}-1,3,4-THIADIAZOL-2-YL)-1-(4-METHYL-1,3-THIAZOL-2-YL)ETHANOL, ENOYL-[ACYL-CARRIER-PROTEIN] REDUCTASE [NADH], NICOTINAMIDE-ADENINE-DINUCLEOTIDE, ... | | Authors: | Read, J.A, Gingell, H, Madhavapeddi, P, Shirude, P.S. | | Deposit date: | 2013-05-31 | | Release date: | 2013-12-11 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (1.89 Å) | | Cite: | Methyl-Thiazoles: A Novel Mode of Inhibition with the Potential to Develop Novel Inhibitors Targeting Inha in Mycobacterium Tuberculosis.
J.Med.Chem., 56, 2013
|
|
7JU8
 
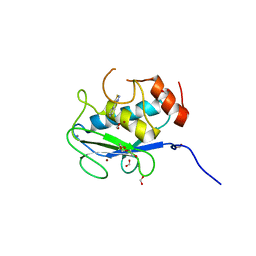 | | X-ray structure of MMP-13 in Complex with 4-(1,2,3-thiadiazol-4-yl)pyridine | | Descriptor: | 4-(1,2,3-thiadiazol-4-yl)pyridine, CALCIUM ION, Collagenase 3, ... | | Authors: | Farrow, N.A. | | Deposit date: | 2020-08-19 | | Release date: | 2021-08-04 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Indole Inhibitors of MMP-13 for Arthritic Disorders
Acs Omega, 6, 2021
|
|
7L28
 
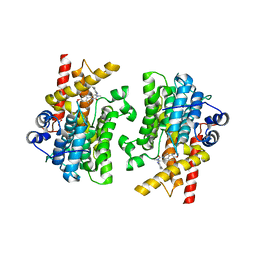 | | Crystal structure of the catalytic domain of human PDE3A bound to Trequinsin | | Descriptor: | (2E)-9,10-dimethoxy-3-methyl-2-[(2,4,6-trimethylphenyl)imino]-2,3,6,7-tetrahydro-4H-pyrimido[6,1-a]isoquinolin-4-one, ACETATE ION, MAGNESIUM ION, ... | | Authors: | Horner, S.W, Garvie, C. | | Deposit date: | 2020-12-16 | | Release date: | 2021-06-16 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Structure of PDE3A-SLFN12 complex reveals requirements for activation of SLFN12 RNase.
Nat Commun, 12, 2021
|
|
7L29
 
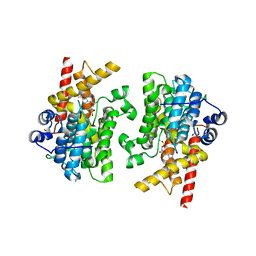 | |
7KWE
 
 | | Crystal structure of the catalytic domain of human PDE3A bound to DNMDP | | Descriptor: | (4~{R})-3-[4-(diethylamino)-3-[oxidanyl(oxidanylidene)-$l^{4}-azanyl]phenyl]-4-methyl-4,5-dihydro-1~{H}-pyridazin-6-one, ACETATE ION, MAGNESIUM ION, ... | | Authors: | Horner, S.W, Garvie, C. | | Deposit date: | 2020-11-30 | | Release date: | 2021-06-16 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Structure of PDE3A-SLFN12 complex reveals requirements for activation of SLFN12 RNase.
Nat Commun, 12, 2021
|
|
7L27
 
 | |
7LRD
 
 | | Cryo-EM of the SLFN12-PDE3A complex: Consensus subset model | | Descriptor: | (4~{R})-3-[4-(diethylamino)-3-[oxidanyl(oxidanylidene)-$l^{4}-azanyl]phenyl]-4-methyl-4,5-dihydro-1~{H}-pyridazin-6-one, MAGNESIUM ION, MANGANESE (II) ION, ... | | Authors: | Fuller, J.R, Garvie, C.W, Lemke, C.T. | | Deposit date: | 2021-02-16 | | Release date: | 2021-06-09 | | Last modified: | 2024-03-06 | | Method: | ELECTRON MICROSCOPY (3.22 Å) | | Cite: | Structure of PDE3A-SLFN12 complex reveals requirements for activation of SLFN12 RNase.
Nat Commun, 12, 2021
|
|
7LRE
 
 | |
7LRC
 
 | | Cryo-EM of the SLFN12-PDE3A complex: PDE3A body refinement | | Descriptor: | (4~{R})-3-[4-(diethylamino)-3-[oxidanyl(oxidanylidene)-$l^{4}-azanyl]phenyl]-4-methyl-4,5-dihydro-1~{H}-pyridazin-6-one, MAGNESIUM ION, MANGANESE (II) ION, ... | | Authors: | Fuller, J.R, Garvie, C.W, Lemke, C.T. | | Deposit date: | 2021-02-16 | | Release date: | 2021-06-09 | | Last modified: | 2024-03-06 | | Method: | ELECTRON MICROSCOPY (2.97 Å) | | Cite: | Structure of PDE3A-SLFN12 complex reveals requirements for activation of SLFN12 RNase.
Nat Commun, 12, 2021
|
|
7LM5
 
 | | Crystal structure of the Zn(II)-bound AdcAII H65A mutant variant of Streptococcus pneumoniae | | Descriptor: | Adhesion protein, CHLORIDE ION, SODIUM ION, ... | | Authors: | Luo, Z, Zupan, M, McDevitt, C.A, Kobe, B. | | Deposit date: | 2021-02-05 | | Release date: | 2021-09-29 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Conformation of the Solute-Binding Protein AdcAII Influences Zinc Uptake in Streptococcus pneumoniae .
Front Cell Infect Microbiol, 11, 2021
|
|
7LM6
 
 | | Crystal structure of the Zn(II)-bound AdcAII H205L mutant variant of Streptococcus pneumoniae | | Descriptor: | Adhesion protein, ZINC ION | | Authors: | Luo, Z, Zupan, M, McDevitt, C.A, Kobe, B. | | Deposit date: | 2021-02-05 | | Release date: | 2021-09-29 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (3.367 Å) | | Cite: | Conformation of the Solute-Binding Protein AdcAII Influences Zinc Uptake in Streptococcus pneumoniae .
Front Cell Infect Microbiol, 11, 2021
|
|
7LM7
 
 | | Crystal structure of the Zn(II)-bound AdcAII E280Q mutant variant of Streptococcus pneumoniae | | Descriptor: | Adhesion protein, CHLORIDE ION, ZINC ION | | Authors: | Luo, Z, Zupan, M, McDevitt, C.A, Kobe, B. | | Deposit date: | 2021-02-05 | | Release date: | 2021-09-29 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (3.12 Å) | | Cite: | Conformation of the Solute-Binding Protein AdcAII Influences Zinc Uptake in Streptococcus pneumoniae .
Front Cell Infect Microbiol, 11, 2021
|
|
