7Y3A
 
 | | Crystal structure of TRIM7 bound to 2C | | Descriptor: | E3 ubiquitin-protein ligase TRIM7,E3 ubiquitin-protein ligase TRIM7,E3 ubiquitin-protein ligase TRIM7,TRIM7-2C | | Authors: | Dong, C, Yan, X. | | Deposit date: | 2022-06-10 | | Release date: | 2022-08-03 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | C-terminal glutamine acts as a C-degron targeted by E3 ubiquitin ligase TRIM7.
Proc.Natl.Acad.Sci.USA, 119, 2022
|
|
7Y3B
 
 | | Crystal structure of TRIM7 bound to GN1 | | Descriptor: | E3 ubiquitin-protein ligase TRIM7,TRIM7-GN1 | | Authors: | Dong, C, Yan, X. | | Deposit date: | 2022-06-10 | | Release date: | 2022-08-03 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (1.76 Å) | | Cite: | C-terminal glutamine acts as a C-degron targeted by E3 ubiquitin ligase TRIM7.
Proc.Natl.Acad.Sci.USA, 119, 2022
|
|
7Y3C
 
 | | Crystal structure of TRIM7 bound to RACO-1 | | Descriptor: | E3 ubiquitin-protein ligase TRIM7,E3 ubiquitin-protein ligase TRIM7,TRIM7-RACO-1 | | Authors: | Dong, C, Yan, X. | | Deposit date: | 2022-06-10 | | Release date: | 2022-08-03 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (1.71 Å) | | Cite: | C-terminal glutamine acts as a C-degron targeted by E3 ubiquitin ligase TRIM7.
Proc.Natl.Acad.Sci.USA, 119, 2022
|
|
5TWY
 
 | | Structure of Maternal Embryonic Leucine Zipper Kinase | | Descriptor: | 2-(benzyloxy)-4-(1H-pyrazol-4-yl)-N-(2,3,4,5-tetrahydro-1H-3-benzazepin-7-yl)benzamide, Maternal embryonic leucine zipper kinase | | Authors: | Seo, H.-S, Dhe-Paganon, S. | | Deposit date: | 2016-11-15 | | Release date: | 2017-11-22 | | Last modified: | 2024-03-06 | | Method: | X-RAY DIFFRACTION (2.91 Å) | | Cite: | MELK is not necessary for the proliferation of basal-like breast cancer cells.
Elife, 6, 2017
|
|
5TWL
 
 | | Structure of Maternal Embryonic Leucine Zipper Kinase | | Descriptor: | 9-(3,5-dichloro-4-hydroxyphenyl)-1-{trans-4-[(dimethylamino)methyl]cyclohexyl}-3,4-dihydropyrimido[5,4-c]quinolin-2(1H)-one, Maternal embryonic leucine zipper kinase | | Authors: | Seo, H.-S, Huang, H.-T, Gray, N.S, Dhe-Paganon, S. | | Deposit date: | 2016-11-14 | | Release date: | 2017-11-22 | | Last modified: | 2024-03-06 | | Method: | X-RAY DIFFRACTION (2.42 Å) | | Cite: | MELK is not necessary for the proliferation of basal-like breast cancer cells.
Elife, 6, 2017
|
|
5TWZ
 
 | | Structure of Maternal Embryonic Leucine Zipper Kinase | | Descriptor: | 7-{[2-methoxy-4-(1H-pyrazol-4-yl)benzoyl]amino}-2,3,4,5-tetrahydro-1H-3-benzazepinium, Maternal embryonic leucine zipper kinase | | Authors: | Seo, H.-Y, Dhe-Paganon, S. | | Deposit date: | 2016-11-15 | | Release date: | 2017-11-22 | | Last modified: | 2024-03-06 | | Method: | X-RAY DIFFRACTION (2.631 Å) | | Cite: | MELK is not necessary for the proliferation of basal-like breast cancer cells.
Elife, 6, 2017
|
|
4GQG
 
 | | Crystal structure of AKR1B10 complexed with NADP+ | | Descriptor: | Aldo-keto reductase family 1 member B10, NADP NICOTINAMIDE-ADENINE-DINUCLEOTIDE PHOSPHATE | | Authors: | Zhang, L, Zheng, X, Chen, S, Zhai, J, Hu, X. | | Deposit date: | 2012-08-23 | | Release date: | 2013-08-28 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (1.92 Å) | | Cite: | Inhibitor selectivity between aldo-keto reductase superfamily members AKR1B10 and AKR1B1: Role of Trp112 (Trp111)
Febs Lett., 587, 2013
|
|
8J49
 
 | | Crystal structure of OY phytoplasma SAP05 in complex with AtSPL5 | | Descriptor: | Sequence-variable mosaic (SVM) signal sequence domain-containing protein, Squamosa promoter-binding-like protein 5, ZINC ION | | Authors: | Dong, C, Yan, X, Yuan, X. | | Deposit date: | 2023-04-19 | | Release date: | 2024-02-28 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (1.66 Å) | | Cite: | Molecular basis of SAP05-mediated ubiquitin-independent proteasomal degradation of transcription factors.
Nat Commun, 15, 2024
|
|
8J4A
 
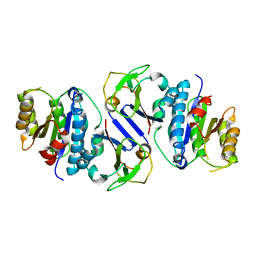 | | Crystal structure of OY phytoplasma SAP05 in complex with AtRPN10 | | Descriptor: | 26S proteasome non-ATPase regulatory subunit 4 homolog, Sequence-variable mosaic (SVM) signal sequence domain-containing protein | | Authors: | Dong, C, Yan, X, Yuan, X. | | Deposit date: | 2023-04-19 | | Release date: | 2024-02-28 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (1.97 Å) | | Cite: | Molecular basis of SAP05-mediated ubiquitin-independent proteasomal degradation of transcription factors.
Nat Commun, 15, 2024
|
|
8J4B
 
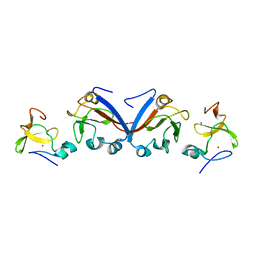 | | Crystal structure of OY phytoplasma SAP05 in complex with AtSPL13 | | Descriptor: | Sequence-variable mosaic (SVM) signal sequence domain-containing protein, Squamosa promoter-binding-like protein 13A, ZINC ION | | Authors: | Dong, C, Yan, X, Yuan, X. | | Deposit date: | 2023-04-19 | | Release date: | 2024-02-28 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Molecular basis of SAP05-mediated ubiquitin-independent proteasomal degradation of transcription factors.
Nat Commun, 15, 2024
|
|
8J48
 
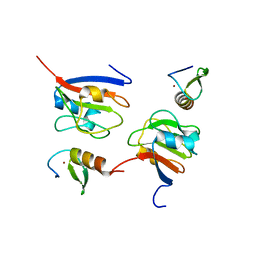 | | Crystal structure of OY phytoplasma SAP05 in complex with AtGATA18 | | Descriptor: | GATA transcription factor 18, Sequence-variable mosaic (SVM) signal sequence domain-containing protein, ZINC ION | | Authors: | Dong, C, Yan, X, Yuan, X. | | Deposit date: | 2023-04-19 | | Release date: | 2024-02-28 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (1.94 Å) | | Cite: | Molecular basis of SAP05-mediated ubiquitin-independent proteasomal degradation of transcription factors.
Nat Commun, 15, 2024
|
|
8TUG
 
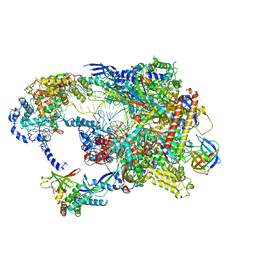 | | Cryo-EM structure of CPD-stalled Pol II in complex with Rad26 (engaged state) | | Descriptor: | DNA (NTS), DNA (TS), DNA repair and recombination protein RAD26, ... | | Authors: | Sarsam, R.D, Lahiri, I, Leschziner, A.E. | | Deposit date: | 2023-08-16 | | Release date: | 2024-01-24 | | Last modified: | 2025-05-14 | | Method: | ELECTRON MICROSCOPY (3.5 Å) | | Cite: | Elf1 promotes Rad26's interaction with lesion-arrested Pol II for transcription-coupled repair.
Proc.Natl.Acad.Sci.USA, 121, 2024
|
|
8QA7
 
 | | Crystal structure of HDAC6 catalytic domain 2 from zebrafish in complex with buffer component. | | Descriptor: | 4-(2-HYDROXYETHYL)-1-PIPERAZINE ETHANESULFONIC ACID, Histone deacetylase 6, POTASSIUM ION, ... | | Authors: | Sandmark, J, Ek, M. | | Deposit date: | 2023-08-22 | | Release date: | 2024-09-11 | | Last modified: | 2025-03-26 | | Method: | X-RAY DIFFRACTION (1.47 Å) | | Cite: | Selective and Bioavailable HDAC6 2-(Difluoromethyl)-1,3,4-oxadiazole Substrate Inhibitors and Modeling of Their Bioactivation Mechanism.
J.Med.Chem., 66, 2023
|
|
8TVP
 
 | | Cryo-EM structure of CPD-stalled Pol II in complex with Rad26 (open state) | | Descriptor: | DNA (NTS), DNA (TS), DNA repair and recombination protein RAD26, ... | | Authors: | Sarsam, R.D, Lahiri, I, Leschziner, A.E. | | Deposit date: | 2023-08-18 | | Release date: | 2024-01-24 | | Last modified: | 2025-05-28 | | Method: | ELECTRON MICROSCOPY (3.7 Å) | | Cite: | Elf1 promotes Rad26's interaction with lesion-arrested Pol II for transcription-coupled repair.
Proc.Natl.Acad.Sci.USA, 121, 2024
|
|
8TVV
 
 | | Cryo-EM structure of backtracked Pol II | | Descriptor: | DNA (NTS), DNA (TS), DNA-directed RNA polymerase II subunit RPB1, ... | | Authors: | Sarsam, R.D, Lahiri, I, Leschziner, A.E. | | Deposit date: | 2023-08-18 | | Release date: | 2024-01-24 | | Last modified: | 2025-05-21 | | Method: | ELECTRON MICROSCOPY (3.7 Å) | | Cite: | Elf1 promotes Rad26's interaction with lesion-arrested Pol II for transcription-coupled repair.
Proc.Natl.Acad.Sci.USA, 121, 2024
|
|
8TVQ
 
 | | Cryo-EM structure of CPD stalled 10-subunit Pol II in complex with Rad26 | | Descriptor: | DNA (NTS), DNA (TS), DNA repair and recombination protein RAD26, ... | | Authors: | Sarsam, R.D, Lahiri, I, Leschziner, A.E. | | Deposit date: | 2023-08-18 | | Release date: | 2024-01-24 | | Last modified: | 2025-05-14 | | Method: | ELECTRON MICROSCOPY (4.6 Å) | | Cite: | Elf1 promotes Rad26's interaction with lesion-arrested Pol II for transcription-coupled repair.
Proc.Natl.Acad.Sci.USA, 121, 2024
|
|
8TVX
 
 | | Cryo-EM structure of CPD-stalled Pol II (Conformation 2) | | Descriptor: | DNA (NTS), DNA (TS), DNA-directed RNA polymerase II subunit RPB1, ... | | Authors: | Sarsam, R.D, Lahiri, I, Leschziner, A.E. | | Deposit date: | 2023-08-18 | | Release date: | 2024-01-24 | | Last modified: | 2025-05-28 | | Method: | ELECTRON MICROSCOPY (3.7 Å) | | Cite: | Elf1 promotes Rad26's interaction with lesion-arrested Pol II for transcription-coupled repair.
Proc.Natl.Acad.Sci.USA, 121, 2024
|
|
8TVW
 
 | | Cryo-EM structure of CPD-stalled Pol II (conformation 1) | | Descriptor: | DNA (NTS), DNA (TS), DNA-directed RNA polymerase II subunit RPB1, ... | | Authors: | Sarsam, R.D, Lahiri, I, Leschziner, A.E. | | Deposit date: | 2023-08-18 | | Release date: | 2024-01-24 | | Last modified: | 2025-05-28 | | Method: | ELECTRON MICROSCOPY (3.6 Å) | | Cite: | Elf1 promotes Rad26's interaction with lesion-arrested Pol II for transcription-coupled repair.
Proc.Natl.Acad.Sci.USA, 121, 2024
|
|
8TVY
 
 | | Cryo-EM structure of CPD lesion containing RNA Polymerase II elongation complex with Rad26 and Elf1 (closed state) | | Descriptor: | DNA (NTS), DNA (TS), DNA-directed RNA polymerase II subunit RPB1, ... | | Authors: | Sarsam, R.D, Lahiri, I, Leschziner, A.E. | | Deposit date: | 2023-08-18 | | Release date: | 2024-01-24 | | Last modified: | 2025-05-28 | | Method: | ELECTRON MICROSCOPY (3.1 Å) | | Cite: | Elf1 promotes Rad26's interaction with lesion-arrested Pol II for transcription-coupled repair.
Proc.Natl.Acad.Sci.USA, 121, 2024
|
|
8TVS
 
 | | Cryo-EM structure of backtracked Pol II in complex with Rad26 | | Descriptor: | DNA (NTS), DNA (TS), DNA repair and recombination protein RAD26, ... | | Authors: | Sarsam, R.D, Lahiri, I, Leschziner, A.E. | | Deposit date: | 2023-08-18 | | Release date: | 2024-01-24 | | Last modified: | 2025-05-28 | | Method: | ELECTRON MICROSCOPY (4.4 Å) | | Cite: | Elf1 promotes Rad26's interaction with lesion-arrested Pol II for transcription-coupled repair.
Proc.Natl.Acad.Sci.USA, 121, 2024
|
|
4LGI
 
 | | N-terminal truncated NleC structure | | Descriptor: | Uncharacterized protein | | Authors: | Li, W.Q, Liu, Y.X, Sheng, X.L, Yan, C.Y, Wang, J.W. | | Deposit date: | 2013-06-28 | | Release date: | 2014-01-15 | | Last modified: | 2024-11-20 | | Method: | X-RAY DIFFRACTION (2.301 Å) | | Cite: | Structure and mechanism of a type III secretion protease, NleC
Acta Crystallogr.,Sect.D, 70, 2014
|
|
8JRE
 
 | | Cryo-EM structure of a designed AAV8-based vector | | Descriptor: | Capsid protein | | Authors: | Ke, X, Luo, S, Zheng, Q, Jiang, H, Liu, F, Sun, X. | | Deposit date: | 2023-06-16 | | Release date: | 2024-04-24 | | Last modified: | 2024-05-15 | | Method: | ELECTRON MICROSCOPY (2.08 Å) | | Cite: | An adeno-associated virus variant enabling efficient ocular-directed gene delivery across species.
Nat Commun, 15, 2024
|
|
4AOI
 
 | | Crystal structure of C-MET kinase domain in complex with 4-(3-((1H- pyrrolo(2,3-b)pyridin-3-yl)methyl)-(1,2,4)triazolo(4,3-b)(1,2,4) triazin-6-yl)benzonitrile | | Descriptor: | 4-[3-(1H-pyrrolo[2,3-b]pyridin-3-ylmethyl)-[1,2,4]triazolo[4,3-b][1,2,4]triazin-6-yl]benzenecarbonitrile, HEPATOCYTE GROWTH FACTOR RECEPTOR | | Authors: | McTigue, M, Grodsky, N, Ryan, K, Cui, J.J. | | Deposit date: | 2012-03-27 | | Release date: | 2012-09-26 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Discovery of a Novel Class of Exquisitely Selective Mesenchymal-Epithelial Transition Factor (C-met) Protein Kinase Inhibitors and Identification of the Clinical Candidate 2-(4-(1-(Quinolin-6-Ylmethyl)-1H-[1,2, 3]Triazolo[4,5-B]Pyrazin-6-Yl)-1H-Pyrazol-1-Yl)Ethanol (Pf-04217903) for the Treatment of Cancer.
J.Med.Chem., 55, 2012
|
|
4AP7
 
 | | Crystal structure of C-MET kinase domain in complex with 4-((6-(4- fluorophenyl)-(1,2,4)triazolo(4,3-b)(1,2,4)triazin-3-yl)methyl)phenol | | Descriptor: | 4-[[6-(4-fluorophenyl)-[1,2,4]triazolo[4,3-b][1,2,4]triazin-3-yl]methyl]phenol, HEPATOCYTE GROWTH FACTOR RECEPTOR | | Authors: | McTigue, M, Wickersham, J. | | Deposit date: | 2012-03-30 | | Release date: | 2012-09-26 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Discovery of a Novel Class of Exquisitely Selective Mesenchymal-Epithelial Transition Factor (C-met) Protein Kinase Inhibitors and Identification of the Clinical Candidate 2-(4-(1-(Quinolin-6-Ylmethyl)-1H-[1,2, 3]Triazolo[4,5-B]Pyrazin-6-Yl)-1H-Pyrazol-1-Yl)Ethanol (Pf-04217903) for the Treatment of Cancer.
J.Med.Chem., 55, 2012
|
|
3FG7
 
 | |
