6O9I
 
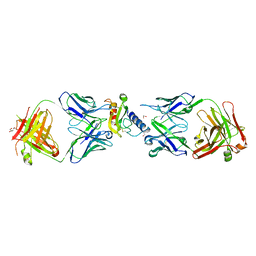 | | Ternary complex of mouse ECD with Fab1 and Fab2 | | Descriptor: | 1,2-ETHANEDIOL, Fab 2 heavy chain, Fab1 heavy chain, ... | | Authors: | Min, X, Wang, Z. | | Deposit date: | 2019-03-13 | | Release date: | 2020-01-22 | | Last modified: | 2024-11-20 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Molecular mechanism of an antagonistic antibody against glucose-dependent insulinotropic polypeptide receptor.
Mabs, 12, 2020
|
|
9IMO
 
 | | Crystal structure of Tubulin-RB3-TTL-Y12 | | Descriptor: | CALCIUM ION, GUANOSINE-5'-DIPHOSPHATE, GUANOSINE-5'-TRIPHOSPHATE, ... | | Authors: | Yan, W, Yang, J.H. | | Deposit date: | 2024-07-04 | | Release date: | 2025-02-26 | | Last modified: | 2025-03-26 | | Method: | X-RAY DIFFRACTION (2.75 Å) | | Cite: | Identification of a ligand-binding site on tubulin mediating the tubulin-RB3 interaction.
Proc.Natl.Acad.Sci.USA, 122, 2025
|
|
9IM5
 
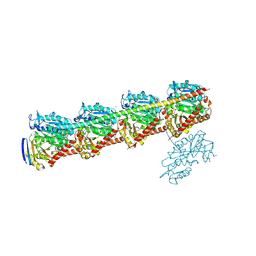 | | Tubulin-RB3(MUT)-TTL-Y12 | | Descriptor: | 2-(N-MORPHOLINO)-ETHANESULFONIC ACID, CALCIUM ION, CHLORIDE ION, ... | | Authors: | Yan, W, Yang, J.H. | | Deposit date: | 2024-07-02 | | Release date: | 2025-02-26 | | Last modified: | 2025-03-26 | | Method: | X-RAY DIFFRACTION (2.86 Å) | | Cite: | Identification of a ligand-binding site on tubulin mediating the tubulin-RB3 interaction.
Proc.Natl.Acad.Sci.USA, 122, 2025
|
|
6VCB
 
 | | Cryo-EM structure of the Glucagon-like peptide-1 receptor in complex with G protein, GLP-1 peptide and a positive allosteric modulator | | Descriptor: | 1-[(1R)-1-(2,6-dichloro-3-methoxyphenyl)ethyl]-6-{2-[(2R)-piperidin-2-yl]phenyl}-1H-benzimidazole, Glucagon-like peptide 1, Glucagon-like peptide 1 receptor, ... | | Authors: | Sun, B, Feng, D, Bueno, A, Kobilka, B, Sloop, K. | | Deposit date: | 2019-12-20 | | Release date: | 2020-07-22 | | Last modified: | 2024-10-23 | | Method: | ELECTRON MICROSCOPY (3.3 Å) | | Cite: | Structural insights into probe-dependent positive allosterism of the GLP-1 receptor.
Nat.Chem.Biol., 16, 2020
|
|
6UMI
 
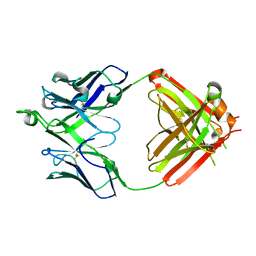 | | Crystal structure of erenumab Fab-b | | Descriptor: | erenumab Fab heavy chain, IgG1, erenumab Fab light chain | | Authors: | Mohr, C. | | Deposit date: | 2019-10-09 | | Release date: | 2020-02-12 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Molecular Insight into Recognition of the CGRPR Complex by Migraine Prevention Therapy Aimovig (Erenumab).
Cell Rep, 30, 2020
|
|
6UMH
 
 | | Crystal structure of erenumab Fab-a | | Descriptor: | (4S)-2-METHYL-2,4-PENTANEDIOL, 1,3-PROPANDIOL, PHOSPHATE ION, ... | | Authors: | Mohr, C. | | Deposit date: | 2019-10-09 | | Release date: | 2020-02-12 | | Last modified: | 2024-10-09 | | Method: | X-RAY DIFFRACTION (2.15 Å) | | Cite: | Molecular Insight into Recognition of the CGRPR Complex by Migraine Prevention Therapy Aimovig (Erenumab).
Cell Rep, 30, 2020
|
|
6UMJ
 
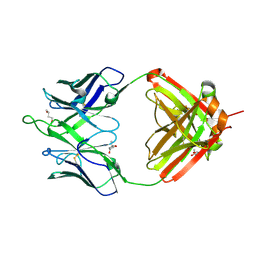 | | Crystal structure of erenumab Fab-c | | Descriptor: | (4S)-2-METHYL-2,4-PENTANEDIOL, 1,3-BUTANEDIOL, erenumab Fab heavy chain, ... | | Authors: | Mohr, C. | | Deposit date: | 2019-10-09 | | Release date: | 2020-02-12 | | Last modified: | 2024-11-13 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | Molecular Insight into Recognition of the CGRPR Complex by Migraine Prevention Therapy Aimovig (Erenumab).
Cell Rep, 30, 2020
|
|
6UMG
 
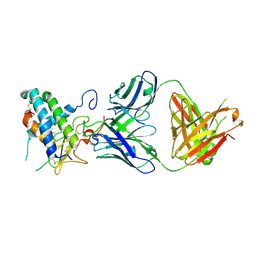 | | Crystal structure of erenumab Fab bound to the extracellular domain of CGRP receptor | | Descriptor: | Calcitonin gene-related peptide type 1 receptor, Receptor activity-modifying protein 1, erenumab Fab heavy chain, ... | | Authors: | Mohr, C. | | Deposit date: | 2019-10-09 | | Release date: | 2020-02-12 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | Molecular Insight into Recognition of the CGRPR Complex by Migraine Prevention Therapy Aimovig (Erenumab).
Cell Rep, 30, 2020
|
|
8AHH
 
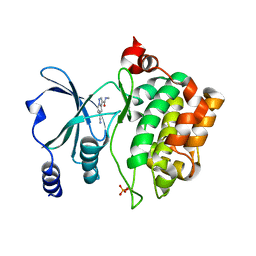 | |
8AHG
 
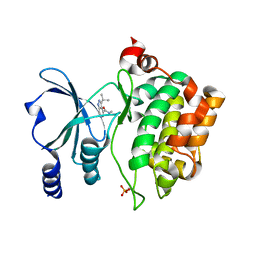 | |
8AHI
 
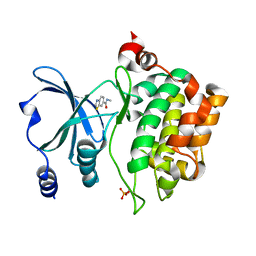 | |
8AHE
 
 | | PAC-FragmentDEL: Photoactivated covalent capture of DNA encoded fragments for hit discovery | | Descriptor: | SULFATE ION, UDP-N-acetylglucosamine 2-epimerase, ~{N},5-dimethyl-1-phenyl-pyrazole-4-sulfonamide | | Authors: | Baker, L.M, Murray, J.B, Hubbard, R.E. | | Deposit date: | 2022-07-21 | | Release date: | 2022-09-14 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (2.108 Å) | | Cite: | PAC-FragmentDEL - photoactivated covalent capture of DNA-encoded fragments for hit discovery.
Rsc Med Chem, 13, 2022
|
|
8AHF
 
 | | PAC-FragmentDEL: Photoactivated covalent capture of DNA encoded fragments for hit discovery | | Descriptor: | (2~{R},4~{S})-4-[bis(fluoranyl)methoxy]-~{N}-methyl-1-(2~{H}-pyrazolo[4,3-b]pyridin-6-ylcarbonyl)pyrrolidine-2-carboxamide, SULFATE ION, UDP-N-acetylglucosamine 2-epimerase | | Authors: | Baker, L.M, Murray, J.B, Hubbard, R.E. | | Deposit date: | 2022-07-21 | | Release date: | 2022-09-14 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (2.271 Å) | | Cite: | PAC-FragmentDEL - photoactivated covalent capture of DNA-encoded fragments for hit discovery.
Rsc Med Chem, 13, 2022
|
|
6LGT
 
 | | Complex structure of HPPD with an inhibitor Y16542 | | Descriptor: | (4S)-2-METHYL-2,4-PENTANEDIOL, 3-(2-chlorophenyl)-6-(1,3-dimethyl-5-oxidanyl-pyrazol-4-yl)carbonyl-1,5-dimethyl-quinazoline-2,4-dione, 4-hydroxyphenylpyruvate dioxygenase, ... | | Authors: | Lin, H.Y, Yang, W.C, Yang, G.F. | | Deposit date: | 2019-12-05 | | Release date: | 2020-10-14 | | Last modified: | 2024-11-13 | | Method: | X-RAY DIFFRACTION (1.794 Å) | | Cite: | Discovery of Novel Pyrazole-Quinazoline-2,4-dione Hybrids as 4-Hydroxyphenylpyruvate Dioxygenase Inhibitors.
J.Agric.Food Chem., 68, 2020
|
|
7EO7
 
 | | Crystal structure of HCoV-NL63 3C-like protease in complex with an inhibitor Shikonin | | Descriptor: | 2-[(1R)-4-methyl-1-oxidanyl-pent-3-enyl]-5,8-bis(oxidanyl)naphthalene-1,4-dione, 3C-like proteinase | | Authors: | Gao, H.X, Zhang, Y.T, Zhou, X.L, Zhong, F.L, Li, J, Zhang, J. | | Deposit date: | 2021-04-21 | | Release date: | 2021-10-20 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2.24916625 Å) | | Cite: | Structure-Based Discovery and Structural Basis of a Novel Broad-Spectrum Natural Product against the Main Protease of Coronavirus.
J.Virol., 96, 2022
|
|
7EO8
 
 | | Crystal structure of SARS coronavirus main protease in complex with an inhibitor Shikonin | | Descriptor: | 2-[(1R)-4-methyl-1-oxidanyl-pent-3-enyl]-5,8-bis(oxidanyl)naphthalene-1,4-dione, 3C-like proteinase | | Authors: | Zhang, Y.T, Gao, H.X, Zhou, H, Zhong, F.L, Hu, X.H, Zhou, X.L, Lin, C, Wang, Q.S, Li, J, Zhang, J. | | Deposit date: | 2021-04-21 | | Release date: | 2021-10-20 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2.2808516 Å) | | Cite: | Structure-Based Discovery and Structural Basis of a Novel Broad-Spectrum Natural Product against the Main Protease of Coronavirus.
J.Virol., 96, 2022
|
|
7DQZ
 
 | | Crystal structure of SARS 3C-like protease in apo form | | Descriptor: | 3C-like proteinase | | Authors: | Zhang, Y.T, Gao, H.X, Zhou, H, Zhong, F.L, Hu, X.H, Zhou, X.L, Lin, C, Wang, Q.S, Li, J, Zhang, J. | | Deposit date: | 2020-12-24 | | Release date: | 2021-09-29 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (1.99 Å) | | Cite: | Structure-Based Discovery and Structural Basis of a Novel Broad-Spectrum Natural Product against the Main Protease of Coronavirus.
J.Virol., 96, 2022
|
|
8DI2
 
 | |
8FPT
 
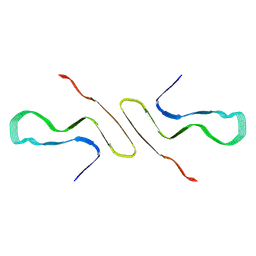 | | STRUCTURE OF ALPHA-SYNUCLEIN FIBRILS DERIVED FROM HUMAN LEWY BODY DEMENTIA TISSUE | | Descriptor: | Alpha-synuclein | | Authors: | Barclay, A.M, Dhavale, D.D, Borcik, C.G, Rau, M.J, Basore, K, Milchberg, M.H, Warmuth, O.A, Kotzbauer, P.T, Rienstra, C.M, Schwieters, C.D. | | Deposit date: | 2023-01-05 | | Release date: | 2023-02-22 | | Last modified: | 2024-12-11 | | Method: | SOLID-STATE NMR | | Cite: | Structure of alpha-synuclein fibrils derived from human Lewy body dementia tissue.
Nat Commun, 15, 2024
|
|
9J7N
 
 | | Cryo-EM structure of TauT | | Descriptor: | BETA-ALANINE, CHLORIDE ION, CHOLESTEROL, ... | | Authors: | Zhao, Y, Xu, H. | | Deposit date: | 2024-08-19 | | Release date: | 2025-04-02 | | Last modified: | 2025-07-02 | | Method: | ELECTRON MICROSCOPY (3.14 Å) | | Cite: | Structural characterization reveals substrate recognition by the taurine transporter TauT.
Cell Discov, 11, 2025
|
|
9J7M
 
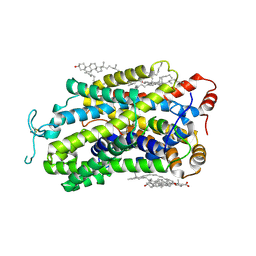 | | Cryo-EM structure of TauT | | Descriptor: | 2-AMINOETHANESULFONIC ACID, CHLORIDE ION, CHOLESTEROL, ... | | Authors: | Zhao, Y, Xu, H. | | Deposit date: | 2024-08-19 | | Release date: | 2025-04-02 | | Last modified: | 2025-05-14 | | Method: | ELECTRON MICROSCOPY (2.82 Å) | | Cite: | Structural characterization reveals substrate recognition by the taurine transporter TauT.
Cell Discov, 11, 2025
|
|
9J7O
 
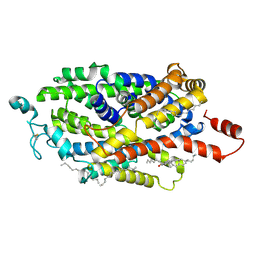 | | Cryo-EM structure of TauT | | Descriptor: | CHLORIDE ION, CHOLESTEROL, HEXADECANE, ... | | Authors: | Zhao, Y, Xu, H. | | Deposit date: | 2024-08-19 | | Release date: | 2025-04-02 | | Last modified: | 2025-05-14 | | Method: | ELECTRON MICROSCOPY (2.77 Å) | | Cite: | Structural characterization reveals substrate recognition by the taurine transporter TauT.
Cell Discov, 11, 2025
|
|
7YE9
 
 | | SARS-CoV-2 Spike (6P) in complex with 3 R1-32 Fabs | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Heavy chain of R1-32 Fab, ... | | Authors: | Liu, B, Gao, X, Li, Z, Chen, X, He, J, Chen, L, Xiong, X. | | Deposit date: | 2022-07-05 | | Release date: | 2022-08-24 | | Last modified: | 2024-11-13 | | Method: | ELECTRON MICROSCOPY (4.17 Å) | | Cite: | SARS-CoV-2 Delta and Omicron variants evade population antibody response by mutations in a single spike epitope.
Nat Microbiol, 7, 2022
|
|
4ZSO
 
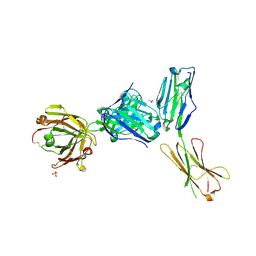 | | Crystal structure of a complex between B7-H6, a tumor cell ligand for natural cytotoxicity receptor NKp30, and an inhibitory antibody | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-[alpha-L-fucopyranose-(1-6)]2-acetamido-2-deoxy-beta-D-glucopyranose, ACETIC ACID, ... | | Authors: | Xu, X, Li, Y, Mariuzza, R.A. | | Deposit date: | 2015-05-13 | | Release date: | 2016-05-18 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Crystal structure of a complex between B7-H6, a tumor cell ligand for natural cytotoxicity receptor NKp30, and an inhibitory antibody
to be published
|
|
7YDY
 
 | | SARS-CoV-2 Spike (6P) in complex with 1 R1-32 Fab | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Heavy chain of R1-32 Fab, ... | | Authors: | Liu, B, Gao, X, Li, Z, Chen, X, He, J, Chen, L, Xiong, X. | | Deposit date: | 2022-07-04 | | Release date: | 2022-08-24 | | Last modified: | 2024-11-13 | | Method: | ELECTRON MICROSCOPY (4.75 Å) | | Cite: | SARS-CoV-2 Delta and Omicron variants evade population antibody response by mutations in a single spike epitope.
Nat Microbiol, 7, 2022
|
|
