4JLQ
 
 | | Crystal structure of human Karyopherin-beta2 bound to the PY-NLS of Saccharomyces cerevisiae NAB2 | | Descriptor: | Nuclear polyadenylated RNA-binding protein NAB2, Transportin-1 | | Authors: | Sampathkumar, P, Gizzi, A, Rout, M.P, Chook, Y.M, Almo, S.C, New York Structural Genomics Research Consortium (NYSGRC), Nucleocytoplasmic Transport: a Target for Cellular Control (NPCXstals) | | Deposit date: | 2013-03-12 | | Release date: | 2013-04-03 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (3.05 Å) | | Cite: | Crystal structure of human Karyopherin beta 2 bound to the PY-NLS of Saccharomyces cerevisiae Nab2.
J.Struct.Funct.Genom., 14, 2013
|
|
4H2H
 
 | | Crystal structure of an enolase (mandalate racemase subgroup, target EFI-502101) from Pelagibaca bermudensis htcc2601, with bound mg and l-4-hydroxyproline betaine (betonicine) | | Descriptor: | (2S,4R)-4-hydroxy-1,1-dimethylpyrrolidinium-2-carboxylate, (4S)-2-METHYL-2,4-PENTANEDIOL, IODIDE ION, ... | | Authors: | Vetting, M.W, Morisco, L.L, Wasserman, S.R, Sojitra, S, Imker, H.J, Gerlt, J.A, Almo, S.C, Enzyme Function Initiative (EFI) | | Deposit date: | 2012-09-12 | | Release date: | 2012-10-10 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Discovery of new enzymes and metabolic pathways by using structure and genome context.
Nature, 502, 2013
|
|
4HB9
 
 | |
2OYC
 
 | | Crystal structure of human pyridoxal phosphate phosphatase | | Descriptor: | Pyridoxal phosphate phosphatase, SODIUM ION, TUNGSTATE(VI)ION | | Authors: | Ramagopal, U.A, Freeman, J, Izuka, M, Toro, R, Sauder, J.M, Burley, S.K, Almo, S.C, New York SGX Research Center for Structural Genomics (NYSGXRC) | | Deposit date: | 2007-02-21 | | Release date: | 2007-03-13 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (1.72 Å) | | Cite: | Structural genomics of protein phosphatases.
J.Struct.Funct.Genom., 8, 2007
|
|
2P69
 
 | | Crystal Structure of Human Pyridoxal Phosphate Phosphatase with PLP | | Descriptor: | CALCIUM ION, PYRIDOXAL-5'-PHOSPHATE, Pyridoxal phosphate phosphatase | | Authors: | Ramagopal, U.A, Freeman, J, Izuka, M, Toro, R, Sauder, J.M, Burley, S.K, Almo, S.C, New York SGX Research Center for Structural Genomics (NYSGXRC) | | Deposit date: | 2007-03-16 | | Release date: | 2007-04-03 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (2.25 Å) | | Cite: | Structural genomics of protein phosphatases.
J.Struct.Funct.Genom., 8, 2007
|
|
2OLA
 
 | | Crystal structure of O-succinylbenzoic acid synthetase from Staphylococcus aureus, cubic crystal form | | Descriptor: | O-succinylbenzoic acid synthetase | | Authors: | Patskovsky, Y, Sauder, J.M, Ozyurt, S, Wasserman, S.R, Smith, D, Dickey, M, Maletic, M, Reyes, C, Gheyi, T, Gerlt, J.A, Almo, S.C, Burley, S.K, New York SGX Research Center for Structural Genomics (NYSGXRC) | | Deposit date: | 2007-01-18 | | Release date: | 2007-02-06 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (2.45 Å) | | Cite: | Loss of quaternary structure is associated with rapid sequence divergence in the OSBS family.
Proc.Natl.Acad.Sci.USA, 111, 2014
|
|
2PGE
 
 | |
7TUX
 
 | | Crystal Structure of Plasmodium falciparum Hypoxanthine-Guanine-Xanthine Phosphoribosyltransferase in complex with [(3S)-4-Hydroxy-3-[({2-amino-4-hydroxy-5H-pyrrolo[3,2-d]pyrimidin-7-yl}methyl)amino]butyl]phosphonic acid | | Descriptor: | 1,2-ETHANEDIOL, ACETIC ACID, Hypoxanthine-guanine-xanthine phosphoribosyltransferase, ... | | Authors: | Harijan, R.K, Minnow, Y.V.T, Bonanno, J.B, Almo, S.C, Schramm, V.L. | | Deposit date: | 2022-02-03 | | Release date: | 2022-12-07 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (1.62 Å) | | Cite: | Inhibition and Mechanism of Plasmodium falciparum Hypoxanthine-Guanine-Xanthine Phosphoribosyltransferase.
Acs Chem.Biol., 17, 2022
|
|
2OKT
 
 | | Crystal structure of O-succinylbenzoic acid synthetase from Staphylococcus aureus, ligand-free form | | Descriptor: | O-succinylbenzoic acid synthetase | | Authors: | Patskovsky, Y, Toro, R, Malashkevich, V, Sauder, J.M, Ozyurt, S, Smith, D, Dickey, M, Maletic, M, Powell, A, Gheyi, T, Wasserman, S.R, Gerlt, J, Burley, S.K, Almo, S.C, New York SGX Research Center for Structural Genomics (NYSGXRC) | | Deposit date: | 2007-01-17 | | Release date: | 2007-01-30 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (1.3 Å) | | Cite: | Loss of quaternary structure is associated with rapid sequence divergence in the OSBS family.
Proc.Natl.Acad.Sci.USA, 111, 2014
|
|
7TWU
 
 | | Crystal structure of human phenylethanolamine N-methyltransferase (PNMT) in complex with (2S)-2-amino-4-(((5-(6-amino-9H-purin-9-yl)-3,4-dihydroxytetrahydrofuran-2-yl)methyl)(4-(7,8-dichloro-1,2,3,4-tetrahydroisoquinolin-4-yl)butyl)amino)butanoic acid and AdoHcy (SAH) | | Descriptor: | 1,2-ETHANEDIOL, 5'-([(3S)-3-amino-3-carboxypropyl]{4-[(4R)-7,8-dichloro-1,2,3,4-tetrahydroisoquinolin-4-yl]butyl}amino)-5'-deoxyadenosine, CADMIUM ION, ... | | Authors: | Harijan, R.K, Mahmoodi, N, Minnow, Y.V.T, Bonanno, J.B, Almo, S.C, Schramm, V.L. | | Deposit date: | 2022-02-07 | | Release date: | 2023-02-15 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Cell-Effective Transition-State Analogue of Phenylethanolamine N -Methyltransferase.
Biochemistry, 62, 2023
|
|
7TX2
 
 | | Crystal structure of human phenylethanolamine N-methyltransferase (PNMT) in complex with (2S)-2-amino-4-(((5-(6-amino-9H-purin-9-yl)-3,4-dihydroxytetrahydrofuran-2-yl)methyl)(4-(7,8-dichloro-1,2,3,4-tetrahydroisoquinolin-4-yl)butyl)amino)butanoic acid | | Descriptor: | 1,2-ETHANEDIOL, 5'-([(3S)-3-amino-3-carboxypropyl]{4-[(4R)-7,8-dichloro-1,2,3,4-tetrahydroisoquinolin-4-yl]butyl}amino)-5'-deoxyadenosine, Phenylethanolamine N-methyltransferase | | Authors: | Harijan, R.K, Mahmoodi, N, Minnow, Y.V.T, Bonanno, J.B, Almo, S.C, Schramm, V.L. | | Deposit date: | 2022-02-07 | | Release date: | 2023-02-15 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (2.43 Å) | | Cite: | Cell-Effective Transition-State Analogue of Phenylethanolamine N -Methyltransferase.
Biochemistry, 62, 2023
|
|
2QJC
 
 | | Crystal structure of a putative diadenosine tetraphosphatase | | Descriptor: | Diadenosine tetraphosphatase, putative, MANGANESE (II) ION, ... | | Authors: | Sugadev, R, Burley, S.K, Swaminathan, S, New York SGX Research Center for Structural Genomics (NYSGXRC) | | Deposit date: | 2007-07-06 | | Release date: | 2007-07-24 | | Last modified: | 2024-11-13 | | Method: | X-RAY DIFFRACTION (2.05 Å) | | Cite: | Structural genomics of protein phosphatases.
J.Struct.Funct.Genom., 8, 2007
|
|
2P27
 
 | | Crystal Structure of Human Pyridoxal Phosphate Phosphatase with Mg2+ at 1.9 A resolution | | Descriptor: | MAGNESIUM ION, Pyridoxal phosphate phosphatase | | Authors: | Ramagopal, U.A, Freeman, J, Izuka, M, Toro, R, Sauder, J.M, Burley, S.K, Almo, S.C, New York SGX Research Center for Structural Genomics (NYSGXRC) | | Deposit date: | 2007-03-07 | | Release date: | 2007-03-13 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Structural genomics of protein phosphatases.
J.Struct.Funct.Genom., 8, 2007
|
|
6AQS
 
 | | Crystal structure of Plasmodium falciparum purine nucleoside phosphorylase (V181D) mutant complexed with DADMe-ImmG and phosphate | | Descriptor: | 1,2-ETHANEDIOL, 2-amino-7-{[(3R,4R)-3-hydroxy-4-(hydroxymethyl)pyrrolidin-1-yl]methyl}-3,5-dihydro-4H-pyrrolo[3,2-d]pyrimidin-4-one, PHOSPHATE ION, ... | | Authors: | Harijan, R.K, Ducati, R.G, Namanja-Magliano, H.A, Bonanno, J.B, Almo, S.C, Schramm, V.L. | | Deposit date: | 2017-08-21 | | Release date: | 2018-02-14 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (1.57 Å) | | Cite: | Genetic resistance to purine nucleoside phosphorylase inhibition in
Proc. Natl. Acad. Sci. U.S.A., 115, 2018
|
|
6AYQ
 
 | | Crystal structure of Campylobacter jejuni 5'-methylthioadenosine/S-adenosyl homocysteine nucleosidase (MTAN) complexed with methylthio-DADMe-Immucillin-A | | Descriptor: | (3R,4S)-1-[(4-AMINO-5H-PYRROLO[3,2-D]PYRIMIDIN-7-YL)METHYL]-4-[(METHYLSULFANYL)METHYL]PYRROLIDIN-3-OL, 5'-methylthioadenosine/S-adenosylhomocysteine nucleosidase, GLYCEROL | | Authors: | Cameron, S.A, Harijan, R.K, Ducati, R.G, Bonanno, J.B, Almo, S.C, Schramm, V.L. | | Deposit date: | 2017-09-08 | | Release date: | 2018-09-12 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (1.42 Å) | | Cite: | Transition-State Analogues of Campylobacter jejuni 5'-Methylthioadenosine Nucleosidase.
ACS Chem. Biol., 13, 2018
|
|
6AYT
 
 | | Crystal structure of Campylobacter jejuni 5'-methylthioadenosine/S-adenosyl homocysteine nucleosidase (MTAN) complexed with pyrazinylthio-DADMe-Immucillin-A | | Descriptor: | (3R,4S)-1-[(4-amino-5H-pyrrolo[3,2-d]pyrimidin-7-yl)methyl]-4-[(pyrazin-2-ylsulfanyl)methyl]pyrrolidin-3-ol, 1,2-ETHANEDIOL, 5'-methylthioadenosine/S-adenosylhomocysteine nucleosidase | | Authors: | Harijan, R.K, Ducati, R.G, Bonanno, J.B, Almo, S.C, Schramm, V.L. | | Deposit date: | 2017-09-08 | | Release date: | 2018-09-12 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (1.85 Å) | | Cite: | Transition-State Analogues of Campylobacter jejuni 5'-Methylthioadenosine Nucleosidase.
ACS Chem. Biol., 13, 2018
|
|
6AYO
 
 | | Crystal structure of Campylobacter jejuni 5'-methylthioadenosine/S-adenosyl homocysteine nucleosidase (MTAN) complexed with 5'-deoxy-5'-Propyl-DADMe-Immucillin-A | | Descriptor: | (3R,4S)-1-[(4-amino-5H-pyrrolo[3,2-d]pyrimidin-7-yl)methyl]-4-propylpyrrolidin-3-ol, 1,2-ETHANEDIOL, 2-[BIS-(2-HYDROXY-ETHYL)-AMINO]-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, ... | | Authors: | Harijan, R.K, Ducati, R.G, Bonanno, J.B, Almo, S.C, Schramm, V.L. | | Deposit date: | 2017-09-08 | | Release date: | 2018-09-12 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (1.67 Å) | | Cite: | Transition-State Analogues of Campylobacter jejuni 5'-Methylthioadenosine Nucleosidase.
ACS Chem. Biol., 13, 2018
|
|
6AYR
 
 | | Crystal structure of Campylobacter jejuni 5'-methylthioadenosine/S-adenosyl homocysteine nucleosidase (MTAN) complexed with butylthio-DADMe-Immucillin-A | | Descriptor: | (3R,4S)-1-[(4-amino-5H-pyrrolo[3,2-d]pyrimidin-7-yl)methyl]-4-[(butylsulfanyl)methyl]pyrrolidin-3-ol, 1,2-ETHANEDIOL, 5'-methylthioadenosine/S-adenosylhomocysteine nucleosidase | | Authors: | Harijan, R.K, Ducati, R.G, Bonanno, J.B, Almo, S.C, Schramm, V.L. | | Deposit date: | 2017-09-08 | | Release date: | 2018-09-12 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (1.95 Å) | | Cite: | Transition-State Analogues of Campylobacter jejuni 5'-Methylthioadenosine Nucleosidase.
ACS Chem. Biol., 13, 2018
|
|
6AYM
 
 | | Crystal structure of Campylobacter jejuni 5'-methylthioadenosine/S-adenosyl homocysteine nucleosidase (MTAN) | | Descriptor: | 1,2-ETHANEDIOL, 5'-methylthioadenosine/S-adenosylhomocysteine nucleosidase | | Authors: | Harijan, R.K, Ducati, R.G, Bonanno, J.B, Almo, S.C, Schramm, V.L. | | Deposit date: | 2017-09-08 | | Release date: | 2018-09-12 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (1.25 Å) | | Cite: | Transition-State Analogues of Campylobacter jejuni 5'-Methylthioadenosine Nucleosidase.
ACS Chem. Biol., 13, 2018
|
|
6AYS
 
 | | Crystal structure of Campylobacter jejuni 5'-methylthioadenosine/S-adenosyl homocysteine nucleosidase (MTAN) complexed with hexylthio-DADMe-Immucillin-A | | Descriptor: | (3R,4S)-1-[(4-amino-5H-pyrrolo[3,2-d]pyrimidin-7-yl)methyl]-4-[(hexylsulfanyl)methyl]pyrrolidin-3-ol, 1,2-ETHANEDIOL, 5'-methylthioadenosine/S-adenosylhomocysteine nucleosidase | | Authors: | Harijan, R.K, Ducati, R.G, Bonanno, J.B, Almo, S.C, Schramm, V.L. | | Deposit date: | 2017-09-08 | | Release date: | 2018-09-12 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Transition-State Analogues of Campylobacter jejuni 5'-Methylthioadenosine Nucleosidase.
ACS Chem. Biol., 13, 2018
|
|
6AQU
 
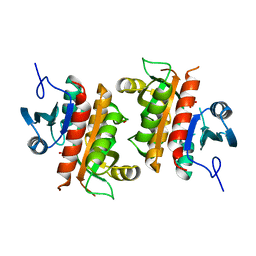 | | Crystal Structure of Plasmodium falciparum purine nucleoside phosphorylase: The M183L mutant | | Descriptor: | PHOSPHATE ION, Purine nucleoside phosphorylase | | Authors: | Harijan, R.K, Ducati, R.G, Namanja-Magliano, H.A, Bonanno, J.B, Almo, S.C, Schramm, V.L. | | Deposit date: | 2017-08-21 | | Release date: | 2018-03-28 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Genetic resistance to purine nucleoside phosphorylase inhibition inPlasmodium falciparum.
Proc. Natl. Acad. Sci. U.S.A., 115, 2018
|
|
5UGF
 
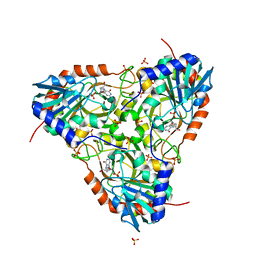 | | Crystal structure of human purine nucleoside phosphorylase (F159Y) mutant complexed with DADMe-ImmG and phosphate | | Descriptor: | 2-amino-7-{[(3R,4R)-3-hydroxy-4-(hydroxymethyl)pyrrolidin-1-yl]methyl}-3,5-dihydro-4H-pyrrolo[3,2-d]pyrimidin-4-one, PHOSPHATE ION, Purine nucleoside phosphorylase | | Authors: | Harijan, R.K, Cameron, S.A, Bonanno, J.B, Almo, S.C, Schramm, V.L. | | Deposit date: | 2017-01-08 | | Release date: | 2017-06-21 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Catalytic-site design for inverse heavy-enzyme isotope effects in human purine nucleoside phosphorylase.
Proc. Natl. Acad. Sci. U.S.A., 114, 2017
|
|
5VJN
 
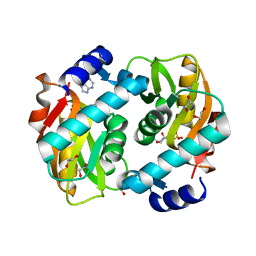 | | Crystal Structure of Adenine Phosphoribosyltransferase from Saccharomyces cerevisiae Complexed with D-2,5-Dideoxy-2,5-Imino-Altritol 1,6-Bisphosphate (D-DIAB) and Adenine | | Descriptor: | 1,2-ETHANEDIOL, ADENINE, Adenine phosphoribosyltransferase 1, ... | | Authors: | Harijan, R.K, Ducati, R.G, Bonanno, J.B, Almo, S.C, Schramm, V.L. | | Deposit date: | 2017-04-19 | | Release date: | 2017-12-27 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (1.78 Å) | | Cite: | Synthesis of bis-Phosphate Iminoaltritol Enantiomers and Structural Characterization with Adenine Phosphoribosyltransferase.
ACS Chem. Biol., 13, 2018
|
|
5VJP
 
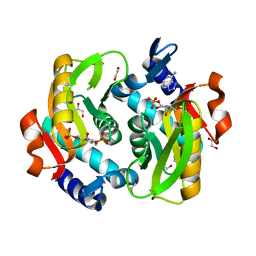 | | Crystal Structure of Adenine Phosphoribosyltransferase from Saccharomyces cerevisiae Complexed with L-2,5-Dideoxy-2,5-Imino-Altritol 1,6-Bisphosphate (L-DIAB) and Adenine | | Descriptor: | 1,2-ETHANEDIOL, ADENINE, Adenine phosphoribosyltransferase 1, ... | | Authors: | Harijan, R.K, Ducati, R.G, Bonanno, J.B, Almo, S.C, Schramm, V.L. | | Deposit date: | 2017-04-19 | | Release date: | 2017-12-27 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (1.98 Å) | | Cite: | Synthesis of bis-Phosphate Iminoaltritol Enantiomers and Structural Characterization with Adenine Phosphoribosyltransferase.
ACS Chem. Biol., 13, 2018
|
|
6DYV
 
 | | Crystal structure of Helicobacter pylori 5'-methylthioadenosine/S-adenosyl homocysteine nucleosidase (MTAN) complexed with (3R,4S)-1-((4-amino-5H-pyrrolo[3,2-d]pyrimidin-7-yl)methyl)-4-((pent-4-yn-1-ylthio)methyl)pyrrolidin-3-ol | | Descriptor: | (3R,4S)-1-[(4-amino-5H-pyrrolo[3,2-d]pyrimidin-7-yl)methyl]-4-{[(pent-4-yn-1-yl)sulfanyl]methyl}pyrrolidin-3-ol, 1,2-ETHANEDIOL, 5'-methylthioadenosine/S-adenosylhomocysteine nucleosidase | | Authors: | Harijan, R.K, Ducati, R.G, Bonanno, J.B, Almo, S.C, Schramm, V.L. | | Deposit date: | 2018-07-02 | | Release date: | 2019-03-20 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (1.62 Å) | | Cite: | Selective Inhibitors of Helicobacter pylori Methylthioadenosine Nucleosidase and Human Methylthioadenosine Phosphorylase.
J. Med. Chem., 62, 2019
|
|
