1G2W
 
 | | E177S MUTANT OF THE PYRIDOXAL-5'-PHOSPHATE ENZYME D-AMINO ACID AMINOTRANSFERASE | | Descriptor: | ACETATE ION, D-ALANINE AMINOTRANSFERASE, PYRIDOXAL-5'-PHOSPHATE | | Authors: | Lepore, B.W, Ringe, D. | | Deposit date: | 2000-10-21 | | Release date: | 2000-11-15 | | Last modified: | 2021-11-03 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Studies on an Active Site Residue, E177, That Affects Binding of the Coenzyme in D-Amino Acid Transaminase, and Mechanistic Studies on a Suicide Substrate
Biochemistry and Molecular Biology of Vitamin B6 and PQQ-dependent Proteins, 10th Annual International Symposium on Vitamin B6, 2000
|
|
4CSK
 
 | | human Aquaporin | | Descriptor: | AQUAPORIN-1 | | Authors: | Ruiz-Carrillo, D, To-Yiu-Ying, J, Darwis, D, Soon, C.H, Cornvik, T, Torres, J, Lescar, J. | | Deposit date: | 2014-03-08 | | Release date: | 2014-12-24 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (3.28 Å) | | Cite: | Crystallization and Preliminary Crystallographic Analysis of Human Aquaporin 1 at a Resolution of 3.28 A.
Acta Crystallogr.,Sect.F, 70, 2014
|
|
4D2E
 
 | | Crystal structure of an integral membrane kinase - v2.3 | | Descriptor: | (2R)-2,3-DIHYDROXYPROPYL(7Z)-PENTADEC-7-ENOATE, (2S)-2,3-DIHYDROXYPROPYL(7Z)-PENTADEC-7-ENOATE, CITRATE ANION, ... | | Authors: | Li, D, Boland, C, Caffrey, M. | | Deposit date: | 2014-05-09 | | Release date: | 2014-07-23 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (2.28 Å) | | Cite: | Cell-Free Expression and in Meso Crystallisation of an Integral Membrane Kinase for Structure Determination.
Cell.Mol.Life Sci., 71, 2014
|
|
1P8T
 
 | | Crystal structure of Nogo-66 Receptor | | Descriptor: | 2-acetamido-2-deoxy-alpha-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose, Reticulon 4 receptor | | Authors: | Barton, W.A, Liu, B.P, Tzvetkova, D, Jeffrey, P.D, Fournier, A.E, Sah, D, Cate, R, Strittmatter, S.M, Nikolov, D.B. | | Deposit date: | 2003-05-07 | | Release date: | 2003-05-20 | | Last modified: | 2024-11-13 | | Method: | X-RAY DIFFRACTION (3.2 Å) | | Cite: | Structure and axon outgrowth inhibitor binding of the Nogo-66 receptor and related proteins
Embo J., 22, 2003
|
|
1GJ4
 
 | | SELECTIVITY AT S1, H2O DISPLACEMENT, UPA, TPA, SER190/ALA190 PROTEASE, STRUCTURE-BASED DRUG DESIGN | | Descriptor: | 6-CHLORO-2-(2-HYDROXY-BIPHENYL-3-YL)-1H-INDOLE-5-CARBOXAMIDINE, ACETYL HIRUDIN, SODIUM ION, ... | | Authors: | Katz, B.A, Sprengeler, P.A, Luong, C, Verner, E, Spencer, J.R, Breitenbucher, J.G, Hui, H, McGee, D, Allen, D, Martelli, A, Mackman, R.L. | | Deposit date: | 2001-04-27 | | Release date: | 2002-04-27 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (1.81 Å) | | Cite: | Engineering inhibitors highly selective for the S1 sites of Ser190 trypsin-like serine protease drug targets.
Chem.Biol., 8, 2001
|
|
1GJD
 
 | | ENGINEERING INHIBITORS HIGHLY SELECTIVE FOR THE S1 SITES OF SER190 TRYPSIN-LIKE SERINE PROTEASE DRUG TARGETS | | Descriptor: | CITRIC ACID, N-(4-CARBAMIMIDOYL-3-CHORO-PHENYL)-2-HYDROXY-3-IODO-5-METHYL-BENZAMIDE, UROKINASE-TYPE PLASMINOGEN ACTIVATOR | | Authors: | Katz, B.A, Sprengeler, P.A, Luong, C, Verner, E, Spencer, J.R, Breitenbucher, J.G, Hui, H, McGee, D, Allen, D, Martelli, A, Mackman, R.L. | | Deposit date: | 2001-05-03 | | Release date: | 2002-05-03 | | Last modified: | 2024-11-20 | | Method: | X-RAY DIFFRACTION (1.75 Å) | | Cite: | Engineering inhibitors highly selective for the S1 sites of Ser190 trypsin-like serine protease drug targets.
Chem.Biol., 8, 2001
|
|
1GJ7
 
 | | ENGINEERING INHIBITORS HIGHLY SELECTIVE FOR THE S1 SITES OF SER190 TRYPSIN-LIKE SERINE PROTEASE DRUG TARGETS | | Descriptor: | 6-CHLORO-2-(2-HYDROXY-BIPHENYL-3-YL)-1H-INDOLE-5-CARBOXAMIDINE, CITRIC ACID, UROKINASE-TYPE PLASMINOGEN ACTIVATOR | | Authors: | Katz, B.A, Sprengeler, P.A, Luong, C, Verner, E, Spencer, J.R, Breitenbucher, J.G, Hui, H, McGee, D, Allen, D, Martelli, A, Mackman, R.L. | | Deposit date: | 2001-04-27 | | Release date: | 2002-04-27 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | Engineering inhibitors highly selective for the S1 sites of Ser190 trypsin-like serine protease drug targets.
Chem.Biol., 8, 2001
|
|
6Z3Z
 
 | |
2OXR
 
 | | PAB0955 crystal structure : a GTPase in GDP and Mg bound form from Pyrococcus abyssi (after GTP hydrolysis) | | Descriptor: | ATP(GTP)binding protein, GUANOSINE-5'-DIPHOSPHATE, MAGNESIUM ION | | Authors: | Gras, S, Carpentier, P, Armengaud, J, Housset, D. | | Deposit date: | 2007-02-21 | | Release date: | 2007-04-24 | | Last modified: | 2024-11-13 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Structural insights into a new homodimeric self-activated GTPase family.
Embo Rep., 8, 2007
|
|
6TZ7
 
 | | Crystal Structure of Aspergillus fumigatus Calcineurin A, Calcineurin B, FKBP12 and FK506 (Tacrolimus) | | Descriptor: | 1,2-ETHANEDIOL, 8-DEETHYL-8-[BUT-3-ENYL]-ASCOMYCIN, CALCIUM ION, ... | | Authors: | Fox III, D, Horanyi, P.S. | | Deposit date: | 2019-08-10 | | Release date: | 2019-09-18 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Harnessing calcineurin-FK506-FKBP12 crystal structures from invasive fungal pathogens to develop antifungal agents.
Nat Commun, 10, 2019
|
|
1QW8
 
 | | Crystal structure of a family 51 alpha-L-arabinofuranosidase in complex with Ara-alpha(1,3)-Xyl | | Descriptor: | Alpha-L-arabinofuranosidase, alpha-L-arabinofuranose-(1-3)-beta-D-xylopyranose | | Authors: | Hoevel, K, Shallom, D, Niefind, K, Belakhov, V, Shoham, G, Bassov, T, Shoham, Y, Schomburg, D. | | Deposit date: | 2003-09-01 | | Release date: | 2003-10-07 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Crystal structure and snapshots along the reaction pathway of a family 51 alpha-L-arabinofuranosidase
Embo J., 22, 2003
|
|
6QMR
 
 | | Complement factor D in complex with the inhibitor (S)-2-(2-((3'-(1-amino-2-hydroxyethyl)-[1,1'-biphenyl]-3-yl)methoxy)phenyl)acetic acid | | Descriptor: | 2-[2-[[3-[3-[(1~{S})-1-azanyl-2-oxidanyl-ethyl]phenyl]phenyl]methoxy]phenyl]ethanoic acid, Complement factor D | | Authors: | Karki, R, Powers, J, Mainolfi, N, Anderson, K, Belanger, D, Liu, D, Jendza, K, Gelin, C.F, Solovay, C, Mac Sweeeny, A, Delgado, O, Crowley, M, Liao, S.-M, Argikar, U.A, Flohr, S, La Bonte, L.R, Lorthiois, E.L, Vulpetti, A, Cumin, F, Brown, A, Adams, C, Jaffee, B, Mogi, M. | | Deposit date: | 2019-02-08 | | Release date: | 2019-04-24 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Design, Synthesis, and Preclinical Characterization of Selective Factor D Inhibitors Targeting the Alternative Complement Pathway.
J.Med.Chem., 62, 2019
|
|
4WYA
 
 | | Crystal structure of 7,8-diaminopelargonic acid synthase (BioA) from Mycobacterium tuberculosis, complexed with a fragment hit | | Descriptor: | 5-(pyridin-2-yl)thiophene-2-carboxamide, Adenosylmethionine-8-amino-7-oxononanoate aminotransferase, PYRIDOXAL-5'-PHOSPHATE | | Authors: | Finzel, B.C, Dai, D, Geders, T.W. | | Deposit date: | 2014-11-17 | | Release date: | 2015-07-15 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Fragment-Based Exploration of Binding Site Flexibility in Mycobacterium tuberculosis BioA.
J.Med.Chem., 58, 2015
|
|
2PE9
 
 | |
6Z4R
 
 | |
6Z4T
 
 | |
1RNW
 
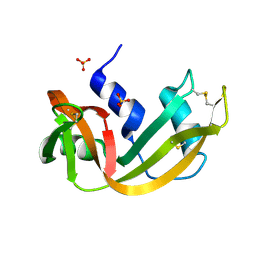 | | RECOMBINANT RIBONUCLEASE A CRYSTALLIZED FROM 80% AMMONIUM SULPHATE | | Descriptor: | RIBONUCLEASE A, SULFATE ION | | Authors: | Fedorov, A.A, Joseph-Mccarthy, D, Fedorov, E.V, Sirakova, D, Graf, I, Almo, S.C. | | Deposit date: | 1996-11-08 | | Release date: | 1997-04-01 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Ionic interactions in crystalline bovine pancreatic ribonuclease A.
Biochemistry, 35, 1996
|
|
4WZN
 
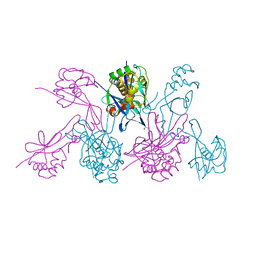 | | CRYSTAL STRUCTURE OF THE 2B PROTEIN SOLUBLE DOMAIN FROM HEPATITIS A VIRUS | | Descriptor: | GLYCEROL, Genome polyprotein | | Authors: | Garriga, D, Vives-Adrian, L, Buxaderas, M, Ferreira-da-Silva, F, Almeida, B, Macedo-Ribeiro, S, Pereira, P.J, Verdaguer, N. | | Deposit date: | 2014-11-20 | | Release date: | 2015-01-28 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | Structural Basis for Host Membrane Remodeling Induced by Protein 2B of Hepatitis A Virus.
J.Virol., 89, 2015
|
|
1R17
 
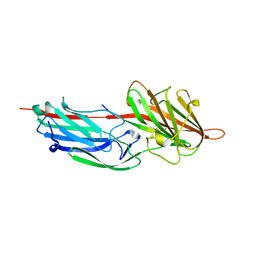 | | Crystal Structure Analysis of S.epidermidis adhesin SdrG binding to Fibrinogen (adhesin-ligand complex) | | Descriptor: | CALCIUM ION, fibrinogen-binding protein SdrG, fibrinopeptide B | | Authors: | Ponnuraj, K, Bowden, M.G, Davis, S, Gurusiddappa, S, Moore, D, Choe, D, Xu, Y, Hook, M, Narayana, S.V.L. | | Deposit date: | 2003-09-23 | | Release date: | 2003-10-28 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (1.86 Å) | | Cite: | A "dock, lock and latch" Structural Model for a Staphylococcal Adhesin Binding to Fibrinogen
Cell(Cambridge,Mass.), 115, 2003
|
|
1R4L
 
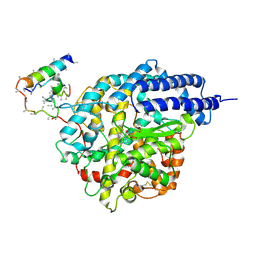 | | Inhibitor Bound Human Angiotensin Converting Enzyme-Related Carboxypeptidase (ACE2) | | Descriptor: | (S,S)-2-{1-CARBOXY-2-[3-(3,5-DICHLORO-BENZYL)-3H-IMIDAZOL-4-YL]-ETHYLAMINO}-4-METHYL-PENTANOIC ACID, 2-acetamido-2-deoxy-beta-D-glucopyranose, CHLORIDE ION, ... | | Authors: | Towler, P, Staker, B, Prasad, S.G, Menon, S, Ryan, D, Tang, J, Parsons, T, Fisher, M, Williams, D, Dales, N.A, Patane, M.A, Pantoliano, M.W. | | Deposit date: | 2003-10-07 | | Release date: | 2004-02-03 | | Last modified: | 2024-11-20 | | Method: | X-RAY DIFFRACTION (3 Å) | | Cite: | ACE2 X-ray structures reveal a large hinge-bending motion important for inhibitor binding and catalysis.
J.Biol.Chem., 279, 2004
|
|
1R61
 
 | | The structure of predicted metal-dependent hydrolase from Bacillus stearothermophilus | | Descriptor: | SULFATE ION, ZINC ION, metal-dependent hydrolase | | Authors: | Maderova, J, Borek, D, Tomchick, D, Joachimiak, A, Collart, F, Otwinowski, Z, Midwest Center for Structural Genomics (MCSG) | | Deposit date: | 2003-10-14 | | Release date: | 2004-03-30 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | The structure of potential metal-dependent hydrolase with cyclase activity
To be Published
|
|
8UAD
 
 | | Cryo-EM structure of prefusion-stabilized influenza B hemagglutinin | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Hemagglutinin HA1 chain, ... | | Authors: | Juraszek, J, Milder, F.J, Yu, X, Blokland, S, Overveld, D, Tamara, S, Bakkers, M.J.G, Rutten, L, Sharma, S, Langedijk, J.P.M. | | Deposit date: | 2023-09-20 | | Release date: | 2024-12-25 | | Last modified: | 2025-05-21 | | Method: | ELECTRON MICROSCOPY (2.77 Å) | | Cite: | Engineering a cleaved, prefusion-stabilized influenza B virus hemagglutinin by identification and locking of all six pH switches.
Pnas Nexus, 3, 2024
|
|
1R7G
 
 | | NMR structure of the membrane anchor domain (1-31) of the nonstructural protein 5A (NS5A) of hepatitis C virus (Minimized average structure, Sample in 100mM DPC) | | Descriptor: | Genome polyprotein | | Authors: | Penin, F, Brass, V, Appel, N, Ramboarina, S, Montserret, R, Ficheux, D, Blum, H.E, Bartenschlager, R, Moradpour, D. | | Deposit date: | 2003-10-21 | | Release date: | 2004-08-10 | | Last modified: | 2024-05-22 | | Method: | SOLUTION NMR | | Cite: | Structure and function of the membrane anchor domain of hepatitis C virus nonstructural protein 5A.
J.Biol.Chem., 279, 2004
|
|
1Q90
 
 | | Structure of the cytochrome b6f (plastohydroquinone : plastocyanin oxidoreductase) from Chlamydomonas reinhardtii | | Descriptor: | 1,2-DI-O-ACYL-3-O-[6-DEOXY-6-SULFO-ALPHA-D-GLUCOPYRANOSYL]-SN-GLYCEROL, 1,2-DISTEAROYL-MONOGALACTOSYL-DIGLYCERIDE, 8-HYDROXY-5,7-DIMETHOXY-3-METHYL-2-TRIDECYL-4H-CHROMEN-4-ONE, ... | | Authors: | Stroebel, D, Choquet, Y, Popot, J.-L, Picot, D. | | Deposit date: | 2003-08-22 | | Release date: | 2003-12-09 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (3.1 Å) | | Cite: | An Atypical Haem in the Cytochrome B6F Complex
Nature, 426, 2003
|
|
7P2N
 
 | | E.coli GyrB24 with inhibitor LSJ38 (EBL2684) | | Descriptor: | 2-[[3,4-bis(chloranyl)-5-methyl-1H-pyrrol-2-yl]carbonylamino]-5-oxidanyl-1,3-benzothiazole-6-carboxylic acid, DNA gyrase subunit B, PHOSPHATE ION | | Authors: | Stevenson, C.E.M, Lawson, D.M, Maxwell, A.M, Henderson, S.R, Kikelj, D, Zega, A, Zidar, N, Ilas, J, Tomasic, T, Masic, L.P. | | Deposit date: | 2021-07-06 | | Release date: | 2022-07-20 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (1.16 Å) | | Cite: | Exploring the 5-Substituted 2-Aminobenzothiazole-Based DNA Gyrase B Inhibitors Active against ESKAPE Pathogens.
Acs Omega, 8, 2023
|
|
