8GXG
 
 | | The crystal structure of SARS-CoV-2 main protease in complex with 14a | | Descriptor: | 3C-like proteinase nsp5, N-[(2S)-3-(4-fluorophenyl)-1-oxidanylidene-1-[[(2S,3S)-3-oxidanyl-4-oxidanylidene-1-[(3S)-2-oxidanylidenepiperidin-3-yl]-4-[(phenylmethyl)amino]butan-2-yl]amino]propan-2-yl]-1-benzofuran-2-carboxamide | | Authors: | Zhao, Y, Zhao, J, Shao, M, Yang, H, Rao, Z. | | Deposit date: | 2022-09-20 | | Release date: | 2023-09-27 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (1.69 Å) | | Cite: | Structure-based design of pan-coronavirus inhibitors targeting host cathepsin L and calpain-1.
Signal Transduct Target Ther, 9, 2024
|
|
8GXH
 
 | | The crystal structure of SARS-CoV-2 main protease in complex with 14b | | Descriptor: | 3C-like proteinase nsp5, N-[(2S)-3-cyclohexyl-1-oxidanylidene-1-[[(2S,3R)-3-oxidanyl-4-oxidanylidene-1-[(3S)-2-oxidanylidenepiperidin-3-yl]-4-[(phenylmethyl)amino]butan-2-yl]amino]propan-2-yl]-1-benzofuran-2-carboxamide | | Authors: | Zhao, Y, Zhao, J, Shao, M, Yang, H, Rao, Z. | | Deposit date: | 2022-09-20 | | Release date: | 2023-09-27 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (1.59 Å) | | Cite: | Structure-based design of pan-coronavirus inhibitors targeting host cathepsin L and calpain-1.
Signal Transduct Target Ther, 9, 2024
|
|
3Q1P
 
 | | Crystal structure of CDP-Chase | | Descriptor: | Phosphohydrolase (MutT/nudix family protein), SULFATE ION | | Authors: | Duong-Ly, K.C, Gabelli, S.B, Amzel, L.M. | | Deposit date: | 2010-12-17 | | Release date: | 2011-07-13 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | The Nudix Hydrolase CDP-Chase, a CDP-Choline Pyrophosphatase, Is an Asymmetric Dimer with Two Distinct Enzymatic Activities.
J.Bacteriol., 193, 2011
|
|
7YLL
 
 | | Crystal structure of TTEDbh | | Descriptor: | DNA polymerase IV, MAGNESIUM ION, PHOSPHATE ION | | Authors: | Yan, X, Tian, L, Gao, H. | | Deposit date: | 2022-07-26 | | Release date: | 2023-12-06 | | Method: | X-RAY DIFFRACTION (2.6000092 Å) | | Cite: | Structure and function of extreme TLS DNA polymerase TTEDbh from Thermoanaerobacter tengcongensis.
Int.J.Biol.Macromol., 253, 2023
|
|
8ILQ
 
 | | Structure of SFTSV Gn-Gc heterodimer | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Envelopment polyprotein | | Authors: | Du, S, Peng, R, Qi, J, Li, C. | | Deposit date: | 2023-03-04 | | Release date: | 2023-09-13 | | Last modified: | 2024-03-27 | | Method: | ELECTRON MICROSCOPY (4.3 Å) | | Cite: | Cryo-EM structure of severe fever with thrombocytopenia syndrome virus.
Nat Commun, 14, 2023
|
|
2O1C
 
 | |
2O5W
 
 | | Structure of the E. coli dihydroneopterin triphosphate pyrophosphohydrolase in complex with Sm+3 and pyrophosphate | | Descriptor: | PYROPHOSPHATE, SAMARIUM (III) ION, SODIUM ION, ... | | Authors: | Gabelli, S.B, Bianchet, M.A, Amzel, L.M. | | Deposit date: | 2006-12-06 | | Release date: | 2007-08-28 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Structure and function of the E. coli dihydroneopterin triphosphate pyrophosphatase: a Nudix enzyme involved in folate biosynthesis.
Structure, 15, 2007
|
|
2O6X
 
 | | Crystal Structure of ProCathepsin L1 from Fasciola hepatica | | Descriptor: | Secreted cathepsin L 1 | | Authors: | Brinen, L.S, Dalton, J.P, Geiger, S, Marion, R, Stack, C.M. | | Deposit date: | 2006-12-08 | | Release date: | 2007-12-18 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (1.4 Å) | | Cite: | Structural and functional relationships in the virulence-associated cathepsin L proteases of the parasitic liver fluke, Fasciola hepatica.
J.Biol.Chem., 283, 2008
|
|
8I4T
 
 | | Structure of the asymmetric unit of SFTSV virion | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Envelopment polyprotein | | Authors: | Du, S, Peng, R, Qi, J, Li, C. | | Deposit date: | 2023-01-21 | | Release date: | 2023-09-13 | | Last modified: | 2024-10-23 | | Method: | ELECTRON MICROSCOPY (5.2 Å) | | Cite: | Cryo-EM structure of severe fever with thrombocytopenia syndrome virus.
Nat Commun, 14, 2023
|
|
7W33
 
 | | The crystal structure of human CtsL in complex with 14a | | Descriptor: | N-[(2S)-3-(4-fluorophenyl)-1-oxidanylidene-1-[[(2R,3S)-3-oxidanyl-4-oxidanylidene-1-[(3S)-2-oxidanylidenepiperidin-3-yl]-4-[(phenylmethyl)amino]butan-2-yl]amino]propan-2-yl]-1-benzofuran-2-carboxamide, Procathepsin L | | Authors: | Zhao, Y, Shao, M, Zhao, J, Yang, H, Rao, Z. | | Deposit date: | 2021-11-25 | | Release date: | 2023-05-31 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (2.39 Å) | | Cite: | Structure-based design of pan-coronavirus inhibitors targeting host cathepsin L and calpain-1.
Signal Transduct Target Ther, 9, 2024
|
|
7W34
 
 | | The crystal structure of human CtsL in complex with 14b | | Descriptor: | N-[(2S)-3-cyclohexyl-1-oxidanylidene-1-[[(2S,3S)-3-oxidanyl-4-oxidanylidene-1-[(3S)-2-oxidanylidenepiperidin-3-yl]-4-[(phenylmethyl)amino]butan-2-yl]amino]propan-2-yl]-1-benzofuran-2-carboxamide, Procathepsin L | | Authors: | Zhao, Y, Shao, M, Zhao, J, Yang, H, Rao, Z. | | Deposit date: | 2021-11-25 | | Release date: | 2023-05-31 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (2.89 Å) | | Cite: | Structure-based design of pan-coronavirus inhibitors targeting host cathepsin L and calpain-1.
Signal Transduct Target Ther, 9, 2024
|
|
7WU5
 
 | | Cryo-EM structure of the adhesion GPCR ADGRF1(H565A/T567A) in complex with miniGi | | Descriptor: | Adhesion G-protein coupled receptor F1, CHOLESTEROL, Guanine nucleotide-binding protein G(I)/G(S)/G(O) subunit gamma-2, ... | | Authors: | Qu, X, Qiu, N, Wang, M, Zhao, Q, Wu, B. | | Deposit date: | 2022-02-05 | | Release date: | 2022-04-27 | | Last modified: | 2024-10-16 | | Method: | ELECTRON MICROSCOPY (3 Å) | | Cite: | Structural basis of tethered agonism of the adhesion GPCRs ADGRD1 and ADGRF1.
Nature, 604, 2022
|
|
7WU4
 
 | | Cryo-EM structure of the adhesion GPCR ADGRF1 in complex with miniGi | | Descriptor: | Adhesion G-protein coupled receptor F1, CHOLESTEROL, Guanine nucleotide-binding protein G(I)/G(S)/G(O) subunit gamma-2, ... | | Authors: | Qu, X, Qiu, N, Wang, M, Zhao, Q, Wu, B. | | Deposit date: | 2022-02-05 | | Release date: | 2022-04-27 | | Last modified: | 2024-10-16 | | Method: | ELECTRON MICROSCOPY (3.4 Å) | | Cite: | Structural basis of tethered agonism of the adhesion GPCRs ADGRD1 and ADGRF1.
Nature, 604, 2022
|
|
7WU3
 
 | | Cryo-EM structure of the adhesion GPCR ADGRF1 in complex with miniGs | | Descriptor: | Adhesion G-protein coupled receptor F1, Guanine nucleotide-binding protein G(I)/G(S)/G(O) subunit gamma-2, Guanine nucleotide-binding protein G(I)/G(S)/G(T) subunit beta-1, ... | | Authors: | Qu, X, Qiu, N, Wang, M, Zhao, Q, Wu, B. | | Deposit date: | 2022-02-05 | | Release date: | 2022-04-27 | | Last modified: | 2022-05-11 | | Method: | ELECTRON MICROSCOPY (3.1 Å) | | Cite: | Structural basis of tethered agonism of the adhesion GPCRs ADGRD1 and ADGRF1.
Nature, 604, 2022
|
|
7WU2
 
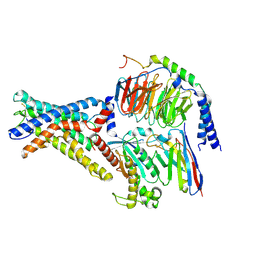 | | Cryo-EM structure of the adhesion GPCR ADGRD1 in complex with miniGs | | Descriptor: | Adhesion G-protein coupled receptor D1, Guanine nucleotide-binding protein G(I)/G(S)/G(O) subunit gamma-2, Guanine nucleotide-binding protein G(I)/G(S)/G(T) subunit beta-1, ... | | Authors: | Qu, X, Qiu, N, Wang, M, Zhao, Q, Wu, B. | | Deposit date: | 2022-02-05 | | Release date: | 2022-04-27 | | Last modified: | 2022-05-11 | | Method: | ELECTRON MICROSCOPY (2.8 Å) | | Cite: | Structural basis of tethered agonism of the adhesion GPCRs ADGRD1 and ADGRF1.
Nature, 604, 2022
|
|
4R82
 
 | | Streptomyces globisporus C-1027 NADH:FAD oxidoreductase SgcE6 in complex with NAD and FAD fragments | | Descriptor: | ACETATE ION, CALCIUM ION, CHLORIDE ION, ... | | Authors: | Tan, K, Bigelow, L, Clancy, S, Babnigg, G, Bingman, C.A, Yennamalli, R, Lohman, J.R, Ma, M, Shen, B, Phillips Jr, G.N, Joachimiak, A, Midwest Center for Structural Genomics (MCSG), Enzyme Discovery for Natural Product Biosynthesis (NatPro) | | Deposit date: | 2014-08-29 | | Release date: | 2014-10-01 | | Last modified: | 2016-11-02 | | Method: | X-RAY DIFFRACTION (1.659 Å) | | Cite: | Crystal Structures of SgcE6 and SgcC, the Two-Component Monooxygenase That Catalyzes Hydroxylation of a Carrier Protein-Tethered Substrate during the Biosynthesis of the Enediyne Antitumor Antibiotic C-1027 in Streptomyces globisporus.
Biochemistry, 55, 2016
|
|
7K8H
 
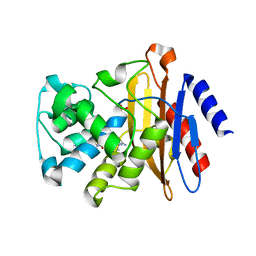 | | Beta-lactamase mixed with Ceftriaxone, 50ms | | Descriptor: | Beta-lactamase, Ceftriaxone, PHOSPHATE ION | | Authors: | Pandey, S, Schmidt, M. | | Deposit date: | 2020-09-27 | | Release date: | 2021-09-22 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (2.60006261 Å) | | Cite: | Observation of substrate diffusion and ligand binding in enzyme crystals using high-repetition-rate mix-and-inject serial crystallography
Iucrj, 8, 2021
|
|
7K8K
 
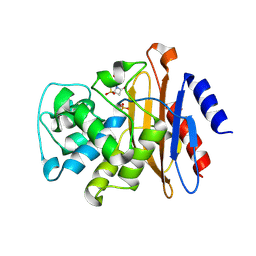 | | Beta-lactamase mixed with Sulbactam, 60ms | | Descriptor: | Beta-lactamase, PHOSPHATE ION, SULBACTAM, ... | | Authors: | Pandey, S, Schmidt, M. | | Deposit date: | 2020-09-27 | | Release date: | 2021-09-22 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | Observation of substrate diffusion and ligand binding in enzyme crystals using high-repetition-rate mix-and-inject serial crystallography
Iucrj, 8, 2021
|
|
7K8L
 
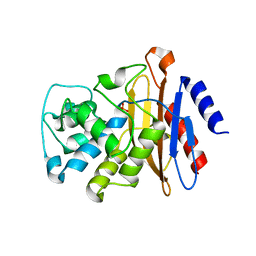 | | Beta-lactamase, Unmixed | | Descriptor: | Beta-lactamase, PHOSPHATE ION | | Authors: | Pandey, S, Schmidt, M. | | Deposit date: | 2020-09-27 | | Release date: | 2021-09-22 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (2.8000102 Å) | | Cite: | Observation of substrate diffusion and ligand binding in enzyme crystals using high-repetition-rate mix-and-inject serial crystallography
Iucrj, 8, 2021
|
|
7E3L
 
 | | Ultrapotent SARS-CoV-2 neutralizing antibodies with protective efficacy against newly emerged mutational variants | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 58G6 heavy chain, 58G6 light chain, ... | | Authors: | Guo, H, Li, T, Liu, F, Gao, Y, Ji, X, Yang, H. | | Deposit date: | 2021-02-09 | | Release date: | 2021-09-15 | | Last modified: | 2024-10-16 | | Method: | ELECTRON MICROSCOPY (3.6 Å) | | Cite: | Potent SARS-CoV-2 neutralizing antibodies with protective efficacy against newly emerged mutational variants.
Nat Commun, 12, 2021
|
|
7E3K
 
 | | Ultrapotent SARS-CoV-2 neutralizing antibodies with protective efficacy against newly emerged mutational variants | | Descriptor: | 13G9 heavy chain, 13G9 light chain, 2-acetamido-2-deoxy-beta-D-glucopyranose, ... | | Authors: | Guo, H, Li, T, Liu, F, Gao, Y, Ji, X, Yang, H. | | Deposit date: | 2021-02-09 | | Release date: | 2021-09-15 | | Last modified: | 2022-03-02 | | Method: | ELECTRON MICROSCOPY (3.9 Å) | | Cite: | Potent SARS-CoV-2 neutralizing antibodies with protective efficacy against newly emerged mutational variants.
Nat Commun, 12, 2021
|
|
7K8E
 
 | | Beta-lactamase mixed with Ceftriaxone, 5ms | | Descriptor: | Beta-lactamase, Ceftriaxone, PHOSPHATE ION | | Authors: | Pandey, S, Schmidt, M. | | Deposit date: | 2020-09-26 | | Release date: | 2021-09-22 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (2.40005636 Å) | | Cite: | Observation of substrate diffusion and ligand binding in enzyme crystals using high-repetition-rate mix-and-inject serial crystallography
Iucrj, 8, 2021
|
|
7K8F
 
 | | Beta-lactamase mixed with Ceftriaxone, 10ms | | Descriptor: | Beta-lactamase, Ceftriaxone, PHOSPHATE ION | | Authors: | Pandey, S, Schmidt, M. | | Deposit date: | 2020-09-26 | | Release date: | 2021-09-22 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (2.60003138 Å) | | Cite: | Observation of substrate diffusion and ligand binding in enzyme crystals using high-repetition-rate mix-and-inject serial crystallography
Iucrj, 8, 2021
|
|
7C2K
 
 | | COVID-19 RNA-dependent RNA polymerase pre-translocated catalytic complex | | Descriptor: | Non-structural protein 7, Non-structural protein 8, RNA (29-MER), ... | | Authors: | Wang, Q, Gao, Y, Ji, W, Mu, A, Rao, Z. | | Deposit date: | 2020-05-07 | | Release date: | 2020-06-03 | | Last modified: | 2024-03-27 | | Method: | ELECTRON MICROSCOPY (2.93 Å) | | Cite: | Structural Basis for RNA Replication by the SARS-CoV-2 Polymerase.
Cell, 182, 2020
|
|
7DGD
 
 | | apo state of class C GPCR | | Descriptor: | Metabotropic glutamate receptor 1 | | Authors: | Zhang, J.Y, Wu, L.J, Luo, F, Hua, T, Liu, Z.J. | | Deposit date: | 2020-11-11 | | Release date: | 2021-09-22 | | Method: | ELECTRON MICROSCOPY (3.96 Å) | | Cite: | Structural insights into the activation initiation of full-length mGlu1.
Protein Cell, 12, 2021
|
|
