7T8R
 
 | |
7TC4
 
 | |
7TB2
 
 | |
7TBT
 
 | |
6DKJ
 
 | | human GIPR ECD and Fab complex | | Descriptor: | 1,2-ETHANEDIOL, Fab heavy chain, Fab light chain, ... | | Authors: | Min, X, Wang, Z. | | Deposit date: | 2018-05-29 | | Release date: | 2019-05-08 | | Method: | X-RAY DIFFRACTION (1.95 Å) | | Cite: | Anti-obesity effects of GIPR antagonists alone and in combination with GLP-1R agonists in preclinical models.
Sci Transl Med, 10, 2018
|
|
7T1D
 
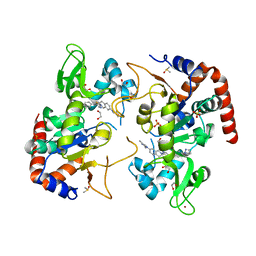 | | Human SIRT2 in complex with small molecule 359 | | Descriptor: | 1,2-ETHANEDIOL, 7-(2,4-dimethyl-1H-imidazol-1-yl)-2-(5-{[4-(1H-pyrazol-1-yl)phenyl]methyl}-1,3-thiazol-2-yl)-1,2,3,4-tetrahydroisoquinoline, DIMETHYL SULFOXIDE, ... | | Authors: | Kulp, J.L, Remiszewski, S, Todd, M, Chiang, L.W. | | Deposit date: | 2021-12-01 | | Release date: | 2023-05-17 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (1.75 Å) | | Cite: | An allosteric inhibitor of sirtuin 2 deacetylase activity exhibits broad-spectrum antiviral activity.
J.Clin.Invest., 133, 2023
|
|
1NVO
 
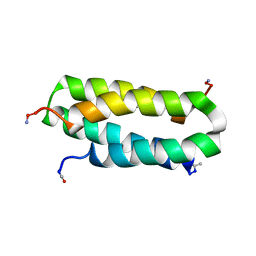 | | Solution structure of a four-helix bundle model, apo-DF1 | | Descriptor: | Homodimeric Alpha2 Four-Helix Bundle | | Authors: | Maglio, O, Nastri, F, Pavone, V, Lombardi, A, DeGrado, W.F. | | Deposit date: | 2003-02-04 | | Release date: | 2003-03-25 | | Last modified: | 2022-02-23 | | Method: | SOLUTION NMR | | Cite: | Preorganization of molecular binding sites in designed diiron proteins
Proc.Natl.Acad.Sci.USA, 100, 2003
|
|
3M4Q
 
 | |
9C7W
 
 | |
8OF2
 
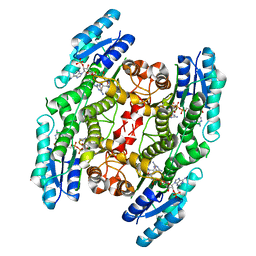 | | Trypanosoma brucei pteridine reductase 1 (TbPTR1) in complex with 2,4,6 triamminopyrimidine (TAP) | | Descriptor: | 1,2-ETHANEDIOL, ACETATE ION, NADPH DIHYDRO-NICOTINAMIDE-ADENINE-DINUCLEOTIDE PHOSPHATE, ... | | Authors: | Tassone, G, Landi, G, Mangani, S, Pozzi, C. | | Deposit date: | 2023-03-13 | | Release date: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (1.48 Å) | | Cite: | The discovery of aryl-2-nitroethyl triamino pyrimidines as anti-Trypanosoma brucei agents.
Eur.J.Med.Chem., 264, 2023
|
|
7XBG
 
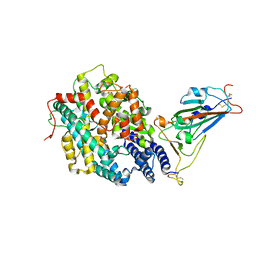 | | The crystal structure of RshSTT182/200 RBD-insert2-T346R-Y496G mutant in complex with human ACE2 | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Processed angiotensin-converting enzyme 2, ... | | Authors: | Hu, Y, Liu, K.F, Han, P, Qi, J.X. | | Deposit date: | 2022-03-21 | | Release date: | 2023-01-11 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (3.37 Å) | | Cite: | Host range and structural analysis of bat-origin RshSTT182/200 coronavirus binding to human ACE2 and its animal orthologs.
Embo J., 42, 2023
|
|
7XBH
 
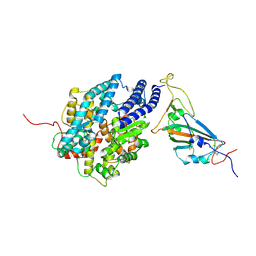 | | The complex structure of RshSTT182/200 RBD bound to human ACE2 | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, Processed angiotensin-converting enzyme 2, RshSTT182/200 coronavirus receptor binding domain, ... | | Authors: | Hu, Y, Liu, K.F, Han, P, Qi, J.X. | | Deposit date: | 2022-03-21 | | Release date: | 2023-01-11 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (3.02 Å) | | Cite: | Host range and structural analysis of bat-origin RshSTT182/200 coronavirus binding to human ACE2 and its animal orthologs.
Embo J., 42, 2023
|
|
7XBF
 
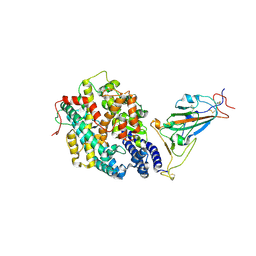 | | The complex structure of RshSTT182/200 RBD-insert2 bound to human ACE2 | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, Processed angiotensin-converting enzyme 2, RshSTT182/200 coronavirus receptor binding domain insert2, ... | | Authors: | Hu, Y, Liu, K.F, Han, P, Qi, J.X. | | Deposit date: | 2022-03-21 | | Release date: | 2023-01-11 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (3.51 Å) | | Cite: | Host range and structural analysis of bat-origin RshSTT182/200 coronavirus binding to human ACE2 and its animal orthologs.
Embo J., 42, 2023
|
|
5MZ3
 
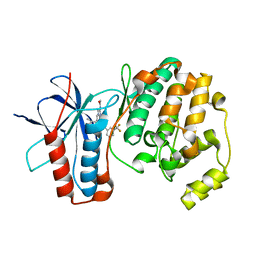 | | P38 ALPHA MUTANT C162S IN COMPLEX WITH CMPD2 [N-(4-Methyl-3-(4,4,5,5-tetramethyl-1,3,2-dioxaborolan-2-yl)phenyl)-3-(trifluoromethyl)benzamide] | | Descriptor: | Mitogen-activated protein kinase 14, ~{N}-[3-(2-acetamidoimidazo[1,2-a]pyridin-6-yl)-4-methyl-phenyl]-3-(trifluoromethyl)benzamide | | Authors: | Cowan-Jacob, S.W, Scheufler, C. | | Deposit date: | 2017-01-30 | | Release date: | 2017-11-15 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (2.15 Å) | | Cite: | Imidazo[1,2-a]pyridin-6-yl-benzamide analogs as potent RAF inhibitors.
Bioorg. Med. Chem. Lett., 27, 2017
|
|
3LZE
 
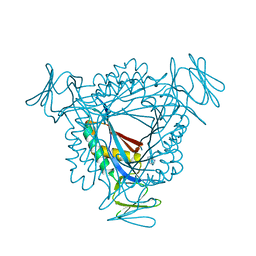 | |
7EN0
 
 | | Structure and Activity of SLAC1 Channels for Stomatal Signaling in Leaves | | Descriptor: | DIUNDECYL PHOSPHATIDYL CHOLINE, SLow Anion Channel 1, SPHINGOSINE | | Authors: | Deng, Y, Kashtoh, H, Wang, Q, Zhen, G, Li, Q, Tang, L, Gao, H, Zhang, C, Qin, L, Su, M, Li, F, Huang, X, Wang, Y, Xie, Q, Clarke, O.B, Hendrickson, W.A, Chen, Y. | | Deposit date: | 2021-04-15 | | Release date: | 2021-05-19 | | Last modified: | 2024-06-05 | | Method: | ELECTRON MICROSCOPY (2.97 Å) | | Cite: | Structure and activity of SLAC1 channels for stomatal signaling in leaves.
Proc.Natl.Acad.Sci.USA, 118, 2021
|
|
3Q01
 
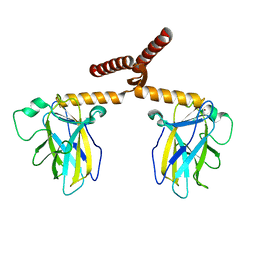 | |
3OQ1
 
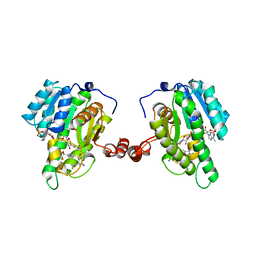 | | Crystal Structure of 11beta-Hydroxysteroid Dehydrogenase-1 (11b-HSD1) in Complex with Diarylsulfone Inhibitor | | Descriptor: | 3-(2-fluoroethyl)-4-({4-[(2S)-1,1,1-trifluoro-2-hydroxypropan-2-yl]phenyl}sulfonyl)benzonitrile, Corticosteroid 11-beta-dehydrogenase isozyme 1, NADP NICOTINAMIDE-ADENINE-DINUCLEOTIDE PHOSPHATE | | Authors: | Wang, Z, Sudom, A, Walker, N.P. | | Deposit date: | 2010-09-02 | | Release date: | 2011-07-20 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | The synthesis and SAR of novel diarylsulfone 11beta-HSD1 inhibitors
Bioorg.Med.Chem.Lett., 20, 2010
|
|
1M1C
 
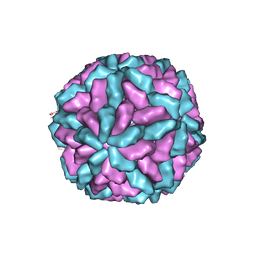 | | Structure of the L-A virus | | Descriptor: | Major coat protein | | Authors: | Naitow, H, Tang, J, Canady, M, Wickner, R.B, Johnson, J.E. | | Deposit date: | 2002-06-18 | | Release date: | 2002-10-02 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (3.5 Å) | | Cite: | L-A virus at 3.4 A resolution reveals particle architecture and mRNA decapping mechanism.
Nat.Struct.Biol., 9, 2002
|
|
3Q05
 
 | |
3Q06
 
 | |
9C80
 
 | | Co-structure of SARS-CoV-2 (COVID-19 with covalent inhibitor | | Descriptor: | (5R,7S,8R)-7-(2-fluorophenyl)-3-[(2-fluorophenyl)carbamoyl]-4,5,6,7-tetrahydropyrazolo[1,5-a]pyrimidine-5-carboxylic acid, 3C-like proteinase nsp5 | | Authors: | Ornelas, E, Knapp, M.S. | | Deposit date: | 2024-06-11 | | Release date: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (1.77 Å) | | Cite: | Identification of Potent, Broad-Spectrum Coronavirus Main Protease Inhibitors for Pandemic Preparedness.
J.Med.Chem., 2024
|
|
9C8Q
 
 | |
8U16
 
 | | The ternary complex structure of DDB1-CRBN-SALL4(ZF1,2)-short bound to Pomalidomide | | Descriptor: | 1,2-ETHANEDIOL, DNA damage-binding protein 1, Protein cereblon, ... | | Authors: | Clifton, M.C, Ma, X, Ornelas, E. | | Deposit date: | 2023-08-30 | | Release date: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (2.9 Å) | | Cite: | Structural and biophysical comparisons of the pomalidomide- and CC-220-induced interactions of SALL4 with cereblon.
Sci Rep, 13, 2023
|
|
8U17
 
 | | The ternary complex structure of DDB1-CRBN-SALL4(ZF1,2)-long bound to Pomalidomide | | Descriptor: | DNA damage-binding protein 1, Protein cereblon, S-Pomalidomide, ... | | Authors: | Clifton, M.C, Ma, X, Ornelas, E. | | Deposit date: | 2023-08-30 | | Release date: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (3.1 Å) | | Cite: | Structural and biophysical comparisons of the pomalidomide- and CC-220-induced interactions of SALL4 with cereblon.
Sci Rep, 13, 2023
|
|
