8H5C
 
 | | Structure of SARS-CoV-2 Omicron BA.2.75 RBD in complex with human ACE2 | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-[alpha-L-fucopyranose-(1-6)]2-acetamido-2-deoxy-beta-D-glucopyranose, ... | | Authors: | Zhao, Z.N, Bai, B, Liu, K.F, Qi, J.X, Gao, G.F. | | Deposit date: | 2022-10-12 | | Release date: | 2023-07-19 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (2.9 Å) | | Cite: | Structural basis for receptor binding and broader interspecies receptor recognition of currently circulating Omicron sub-variants.
Nat Commun, 14, 2023
|
|
6OC9
 
 | | S8 phosphorylated beta amyloid 40 fibrils | | Descriptor: | Amyloid-beta precursor protein, PHOSPHONATE | | Authors: | Qiang, W, Hu, Z.W. | | Deposit date: | 2019-03-22 | | Release date: | 2019-06-05 | | Last modified: | 2024-05-01 | | Method: | SOLID-STATE NMR | | Cite: | Molecular structure of an N-terminal phosphorylated beta-amyloid fibril.
Proc.Natl.Acad.Sci.USA, 116, 2019
|
|
6OIP
 
 | | Crystal structure of MYST acetyltransferase domain in complex with inhibitor 34 | | Descriptor: | 2-fluoro-3-methyl-N'-[(naphthalen-2-yl)sulfonyl]benzohydrazide, GLYCEROL, Histone acetyltransferase KAT8, ... | | Authors: | Hermans, S.J, Chung, M.C, Parker, M.W, Thomas, T, Baell, J.B. | | Deposit date: | 2019-04-09 | | Release date: | 2019-07-03 | | Last modified: | 2024-11-20 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Discovery of Benzoylsulfonohydrazides as Potent Inhibitors of the Histone Acetyltransferase KAT6A.
J.Med.Chem., 62, 2019
|
|
6OIQ
 
 | | Crystal structure of MYST acetyltransferase domain in complex with inhibitor 63 | | Descriptor: | 2-fluoro-N'-(phenylsulfonyl)[1,1'-biphenyl]-3-carbohydrazide, GLYCEROL, Histone acetyltransferase KAT8, ... | | Authors: | Hermans, S.J, Chung, M.C, Parker, M.W, Thomas, T, Baell, J.B. | | Deposit date: | 2019-04-09 | | Release date: | 2019-07-03 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (1.75 Å) | | Cite: | Discovery of Benzoylsulfonohydrazides as Potent Inhibitors of the Histone Acetyltransferase KAT6A.
J.Med.Chem., 62, 2019
|
|
6OIO
 
 | | Crystal structure of MYST acetyltransferase domain in complex with inhibitor 60 | | Descriptor: | Histone acetyltransferase KAT8, N'-(phenylsulfonyl)[1,1'-biphenyl]-3-carbohydrazide, ZINC ION | | Authors: | Hermans, S.J, Chung, M.C, Parker, M.W, Thomas, T, Baell, J.B. | | Deposit date: | 2019-04-09 | | Release date: | 2019-07-03 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Discovery of Benzoylsulfonohydrazides as Potent Inhibitors of the Histone Acetyltransferase KAT6A.
J.Med.Chem., 62, 2019
|
|
6OWI
 
 | | Crystal structure of MYST acetyltransferase domain in complex with inhibitor 85 | | Descriptor: | GLYCEROL, Histone acetyltransferase KAT8, N'-[(2-fluorophenyl)sulfonyl]benzohydrazide, ... | | Authors: | Hermans, S.J, Chung, M.C, Parker, M.W, Thomas, T, Baell, J.B. | | Deposit date: | 2019-05-09 | | Release date: | 2020-04-01 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (1.75 Å) | | Cite: | Discovery of Acylsulfonohydrazide-Derived Inhibitors of the Lysine Acetyltransferase, KAT6A, as Potent Senescence-Inducing Anti-Cancer Agents.
J.Med.Chem., 63, 2020
|
|
5UFI
 
 | | DCN1 bound to DI-591 | | Descriptor: | DCN1-like protein 1, N-[(1S)-1-cyclohexyl-2-{[3-(morpholin-4-yl)propanoyl]amino}ethyl]-N~2~-propanoyl-3-[6-(propan-2-yl)-1,3-benzothiazol-2-yl]-L-alaninamide | | Authors: | Stuckey, J. | | Deposit date: | 2017-01-04 | | Release date: | 2017-11-01 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (2.58 Å) | | Cite: | A potent small-molecule inhibitor of the DCN1-UBC12 interaction that selectively blocks cullin 3 neddylation.
Nat Commun, 8, 2017
|
|
5U18
 
 | | Crystal structure of a methyltransferase involved in the biosynthesis of gentamicin in complex with the Geneticin | | Descriptor: | GENETICIN, N-3'' methyltransferase, S-ADENOSYL-L-HOMOCYSTEINE | | Authors: | Bury, P, Huang, F, Leadlay, P, Dias, M.V.B. | | Deposit date: | 2016-11-28 | | Release date: | 2017-11-01 | | Last modified: | 2024-03-06 | | Method: | X-RAY DIFFRACTION (2.195 Å) | | Cite: | Structural Basis of the Selectivity of GenN, an Aminoglycoside N-Methyltransferase Involved in Gentamicin Biosynthesis.
ACS Chem. Biol., 12, 2017
|
|
5U1E
 
 | | Crystal structure of a methyltransferase involved in the biosynthesis of gentamicin in complex with the kanamycin B | | Descriptor: | (1R,2S,3S,4R,6S)-4,6-DIAMINO-3-[(3-AMINO-3-DEOXY-ALPHA-D-GLUCOPYRANOSYL)OXY]-2-HYDROXYCYCLOHEXYL 2,6-DIAMINO-2,6-DIDEOXY-ALPHA-D-GLUCOPYRANOSIDE, Putative gentamicin methyltransferase, S-ADENOSYL-L-HOMOCYSTEINE | | Authors: | Bury, P, Huang, F, Leadlay, P, Dias, M.V.B. | | Deposit date: | 2016-11-28 | | Release date: | 2017-11-01 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (2.213 Å) | | Cite: | Structural Basis of the Selectivity of GenN, an Aminoglycoside N-Methyltransferase Involved in Gentamicin Biosynthesis.
ACS Chem. Biol., 12, 2017
|
|
5TYQ
 
 | | Crystal structure of a holoenzyme methyltransferase involved in the biosynthesis of gentamicin | | Descriptor: | MAGNESIUM ION, Putative gentamicin methyltransferase, S-ADENOSYL-L-HOMOCYSTEINE | | Authors: | Bury, P, Huang, F, Leadlay, P, Dias, M.V.B. | | Deposit date: | 2016-11-21 | | Release date: | 2017-11-01 | | Last modified: | 2024-03-06 | | Method: | X-RAY DIFFRACTION (2.163 Å) | | Cite: | Structural Basis of the Selectivity of GenN, an Aminoglycoside N-Methyltransferase Involved in Gentamicin Biosynthesis.
ACS Chem. Biol., 12, 2017
|
|
5U4T
 
 | | Crystal structure of a methyltransferase involved in the biosynthesis of gentamicin | | Descriptor: | 2-AMINO-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, IODIDE ION, Putative gentamicin methyltransferase, ... | | Authors: | Bury, P, Huang, F, Leadlay, P, Dias, M.V.B. | | Deposit date: | 2016-12-06 | | Release date: | 2017-11-01 | | Last modified: | 2024-03-06 | | Method: | X-RAY DIFFRACTION (2.086 Å) | | Cite: | Structural Basis of the Selectivity of GenN, an Aminoglycoside N-Methyltransferase Involved in Gentamicin Biosynthesis.
ACS Chem. Biol., 12, 2017
|
|
5U0T
 
 | | Crystal structure of a methyltransferase involved in the biosynthesis of gentamicin in complex with (1R,2S,3S,4R,6S)-4,6-diamino-3-{[3-deoxy-3-(methylamino)-alpha-D-glucopyranosyl]oxy}-2-hydroxycyclohexyl 2,6-diamino-2,6-dideoxy-alpha-D-glucopyranoside | | Descriptor: | (1R,2S,3S,4R,6S)-4,6-diamino-3-{[3-deoxy-3-(methylamino)-alpha-D-glucopyranosyl]oxy}-2-hydroxycyclohexyl 2,6-diamino-2,6-dideoxy-alpha-D-glucopyranoside, MAGNESIUM ION, Putative gentamicin methyltransferase, ... | | Authors: | Bury, P, Huang, F, Leadlay, P, Dias, M.V.B. | | Deposit date: | 2016-11-27 | | Release date: | 2017-11-01 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (2.109 Å) | | Cite: | Structural Basis of the Selectivity of GenN, an Aminoglycoside N-Methyltransferase Involved in Gentamicin Biosynthesis.
ACS Chem. Biol., 12, 2017
|
|
5U19
 
 | | Crystal structure of a methyltransferase involved in the biosynthesis of gentamicin in complex with (1R,2S,3S,4R,6R)-4,6-diamino-3-{[3-deoxy-4-C-methyl-3-(methylamino)-beta-L-arabinopyranosyl]oxy}-2-hydroxycyclohexyl 2-amino-2-deoxy-alpha-D-glucopyranoside | | Descriptor: | (1R,2S,3S,4R,6S)-4,6-diamino-3-{[3-deoxy-4-C-methyl-3-(methylamino)-beta-L-arabinopyranosyl]oxy}-2-hydroxycyclohexyl 2-amino-2-deoxy-alpha-D-glucopyranoside, MAGNESIUM ION, Putative gentamicin methyltransferase, ... | | Authors: | Bury, P, Huang, F, Leadlay, P, Dias, M.V.B. | | Deposit date: | 2016-11-28 | | Release date: | 2017-11-01 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (1.902 Å) | | Cite: | Structural Basis of the Selectivity of GenN, an Aminoglycoside N-Methyltransferase Involved in Gentamicin Biosynthesis.
ACS Chem. Biol., 12, 2017
|
|
5U1I
 
 | | Crystal structure of a methyltransferase involved in the biosynthesis of gentamicin in complex with the methylated Kanamycin B | | Descriptor: | (1R,2S,3S,4R,6S)-4,6-diamino-3-{[3-deoxy-3-(methylamino)-alpha-D-glucopyranosyl]oxy}-2-hydroxycyclohexyl 2,6-diamino-2,6-dideoxy-alpha-D-glucopyranoside, CALCIUM ION, Putative gentamicin methyltransferase, ... | | Authors: | Bury, P, Huang, F, Leadlay, P, Dias, M.V.B. | | Deposit date: | 2016-11-28 | | Release date: | 2017-11-01 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (1.932 Å) | | Cite: | Structural Basis of the Selectivity of GenN, an Aminoglycoside N-Methyltransferase Involved in Gentamicin Biosynthesis.
ACS Chem. Biol., 12, 2017
|
|
5H19
 
 | | EED in complex with PRC2 allosteric inhibitor EED162 | | Descriptor: | 5-(furan-2-ylmethylamino)-9-(phenylmethyl)-8,10-dihydro-7H-[1,2,4]triazolo[3,4-a][2,7]naphthyridine-6-carbonitrile, Histone-lysine N-methyltransferase EZH2, Polycomb protein EED | | Authors: | Zhao, K, Zhao, M, Luo, X, Zhang, H. | | Deposit date: | 2016-10-08 | | Release date: | 2017-01-25 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Discovery and Molecular Basis of a Diverse Set of Polycomb Repressive Complex 2 Inhibitors Recognition by EED
PLoS ONE, 12, 2017
|
|
7RNO
 
 | | Model of the Ac-6-FP/hpMR1/bB2m/TAPBPR complex from integrated docking, NMR and restrained MD | | Descriptor: | Beta-2-microglobulin, Major histocompatibility complex class I-related gene protein, N-(6-formyl-4-oxo-3,4-dihydropteridin-2-yl)acetamide, ... | | Authors: | McShan, A.C, Sgourakis, N.G. | | Deposit date: | 2021-07-29 | | Release date: | 2022-05-11 | | Last modified: | 2022-08-10 | | Method: | SOLUTION NMR | | Cite: | TAPBPR employs a ligand-independent docking mechanism to chaperone MR1 molecules.
Nat.Chem.Biol., 18, 2022
|
|
6ADR
 
 | |
6ADL
 
 | |
6ADS
 
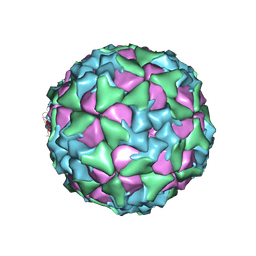 | | Structure of Seneca Valley Virus in acidic conditions | | Descriptor: | VP1, VP2, VP3, ... | | Authors: | Lou, Z.Y, Cao, L. | | Deposit date: | 2018-08-02 | | Release date: | 2019-02-06 | | Last modified: | 2025-06-25 | | Method: | ELECTRON MICROSCOPY (2.84 Å) | | Cite: | Seneca Valley virus attachment and uncoating mediated by its receptor anthrax toxin receptor 1
Proc. Natl. Acad. Sci. U.S.A., 115, 2018
|
|
6ADQ
 
 | | Respiratory Complex CIII2CIV2SOD2 from Mycobacterium smegmatis | | Descriptor: | (2R)-2-(hexadecanoyloxy)-3-{[(S)-hydroxy{[(1R,2R,3R,4R,5R,6S)-2,3,4,5,6-pentahydroxycyclohexyl]oxy}phosphoryl]oxy}propyl (9S)-9-methyloctadecanoate, (2R)-3-(((2-aminoethoxy)(hydroxy)phosphoryl)oxy)-2-(palmitoyloxy)propyl (E)-octadec-9-enoate, (2S)-1-(hexadecanoyloxy)propan-2-yl (10S)-10-methyloctadecanoate, ... | | Authors: | Gong, H.R, Xu, A, Gao, R.G, Ji, W.X, Wang, S.H, Wang, Q, Li, J, Rao, Z.H. | | Deposit date: | 2018-08-01 | | Release date: | 2018-11-14 | | Last modified: | 2025-04-09 | | Method: | ELECTRON MICROSCOPY (3.5 Å) | | Cite: | An electron transfer path connects subunits of a mycobacterial respiratory supercomplex.
Science, 362, 2018
|
|
6ADT
 
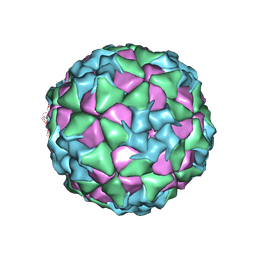 | | Structure of Seneca Valley Virus in neutral condition | | Descriptor: | VP1, VP2, VP3, ... | | Authors: | Lou, Z.Y, Cao, L. | | Deposit date: | 2018-08-02 | | Release date: | 2019-02-06 | | Last modified: | 2025-06-18 | | Method: | ELECTRON MICROSCOPY (3.22 Å) | | Cite: | Seneca Valley virus attachment and uncoating mediated by its receptor anthrax toxin receptor 1
Proc. Natl. Acad. Sci. U.S.A., 115, 2018
|
|
6ADM
 
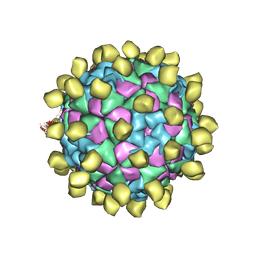 | |
5XYZ
 
 | | The structure of human BTK kinase domain in complex with a covalent inhibitor | | Descriptor: | N-[3-(5-{[(2-chloro-6-fluorophenyl)methyl]amino}-1H-1,2,4-triazol-3-yl)phenyl]propanamide, Tyrosine-protein kinase BTK | | Authors: | Wang, Y.L, Sun, Y.Z, Cao, R, Liu, D, Xie, Y.T, Li, L, Qi, X.B, Huang, N. | | Deposit date: | 2017-07-11 | | Release date: | 2018-05-23 | | Last modified: | 2024-11-20 | | Method: | X-RAY DIFFRACTION (2.64 Å) | | Cite: | In Silico Identification of a Novel Hinge-Binding Scaffold for Kinase Inhibitor Discovery.
J. Med. Chem., 60, 2017
|
|
5XYX
 
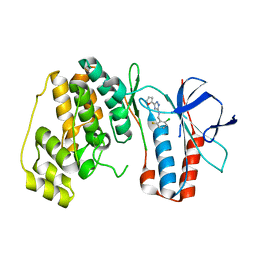 | | The structure of p38 alpha in complex with a triazol inhibitor | | Descriptor: | Mitogen-activated protein kinase 14, N-(2-chloro-6-fluorobenzyl)-5-(furan-2-yl)-2H-1,2,4-triazol-3-amine | | Authors: | Wang, Y.L, Sun, Y.Z, Cao, R, Liu, D, Li, L, Qi, X.B, Huang, N. | | Deposit date: | 2017-07-11 | | Release date: | 2018-01-17 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (2.61 Å) | | Cite: | In Silico Identification of a Novel Hinge-Binding Scaffold for Kinase Inhibitor Discovery.
J. Med. Chem., 60, 2017
|
|
3TWJ
 
 | | Rho-associated protein kinase 1 (ROCK 1) IN COMPLEX WITH RKI1447 | | Descriptor: | 1,2-ETHANEDIOL, 1-[(3-hydroxyphenyl)methyl]-3-(4-pyridin-4-yl-1,3-thiazol-2-yl)urea, Rho-associated protein kinase 1 | | Authors: | Martin, M.P, Zhu, J.-Y, Schonbrunn, E. | | Deposit date: | 2011-09-21 | | Release date: | 2012-08-22 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (2.9 Å) | | Cite: | RKI-1447 Is a Potent Inhibitor of the Rho-Associated ROCK Kinases with Anti-Invasive and Antitumor Activities in Breast Cancer.
Cancer Res., 72, 2012
|
|
