1AXL
 
 | | SOLUTION CONFORMATION OF THE (-)-TRANS-ANTI-[BP]DG ADDUCT OPPOSITE A DELETION SITE IN DNA DUPLEX D(CCATC-[BP]G-CTACC)D(GGTAG--GATGG), NMR, 6 STRUCTURES | | Descriptor: | 1,2,3-TRIHYDROXY-1,2,3,4-TETRAHYDROBENZO[A]PYRENE, DNA DUPLEX D(CCATC-[BP]G-CTACC)D(GGTAG--GATGG) | | Authors: | Feng, B, Gorin, A.A, Kolbanovskiy, A, Hingerty, B.E, Geacintov, N.E, Broyde, S, Patel, D.J. | | Deposit date: | 1997-10-16 | | Release date: | 1998-07-01 | | Last modified: | 2024-05-22 | | Method: | SOLUTION NMR | | Cite: | Solution conformation of the (-)-trans-anti-[BP]dG adduct opposite a deletion site in a DNA duplex: intercalation of the covalently attached benzo[a]pyrene into the helix with base displacement of the modified deoxyguanosine into the minor groove.
Biochemistry, 36, 1997
|
|
1AX7
 
 | | SOLUTION STRUCTURE OF THE [AF]-C8-DG ADDUCT POSITIONED AT A TEMPLATE-PRIMER JUNCTION, NMR, 6 STRUCTURES | | Descriptor: | 2-AMINOFLUORENE, DNA DUPLEX D(AAC-[AF]G-CTACCATCC)D(GGATGGTAG) | | Authors: | Mao, B, Gu, Z, Gorin, A.A, Hingerty, B.E, Broyde, S, Patel, D.J. | | Deposit date: | 1997-10-30 | | Release date: | 1998-07-01 | | Last modified: | 2024-05-22 | | Method: | SOLUTION NMR | | Cite: | Solution structure of the aminofluorene-stacked conformer of the syn [AF]-C8-dG adduct positioned at a template-primer junction.
Biochemistry, 36, 1997
|
|
6O7E
 
 | |
1AXO
 
 | | STRUCTURAL ALIGNMENT OF THE (+)-TRANS-ANTI-[BP]DG ADDUCT POSITIONED OPPOSITE DC AT A DNA TEMPLATE-PRIMER JUNCTION, NMR, 6 STRUCTURES | | Descriptor: | 1,2,3-TRIHYDROXY-1,2,3,4-TETRAHYDROBENZO[A]PYRENE, DNA DUPLEX D(AAC-[BP]G-CTACCATCC)D(GGATGGTAGC) | | Authors: | Feng, B, Gorin, A.A, Hingerty, B.E, Geacintov, N.E, Broyde, S, Patel, D.J. | | Deposit date: | 1997-10-16 | | Release date: | 1998-07-01 | | Last modified: | 2024-05-22 | | Method: | SOLUTION NMR | | Cite: | Structural alignment of the (+)-trans-anti-benzo[a]pyrene-dG adduct positioned opposite dC at a DNA template-primer junction.
Biochemistry, 36, 1997
|
|
1B5K
 
 | | 3,N4-ETHENO-2'-DEOXYCYTIDINE OPPOSITE THYMIDINE IN AN 11-MER DUPLEX, SOLUTION STRUCTURE FROM NMR AND MOLECULAR DYNAMICS | | Descriptor: | DNA (5'-D(*CP*GP*TP*AP*CP*EDCP*CP*AP*TP*GP*C)-3'), DNA (5'-D(*GP*CP*AP*TP*GP*TP*GP*TP*AP*CP*G)-3') | | Authors: | Cullinan, D, Korobka, A, Grollman, A.P, Patel, D.J, Eisenberg, M, De Santos, C.L. | | Deposit date: | 1999-01-07 | | Release date: | 1999-01-13 | | Last modified: | 2023-12-27 | | Method: | SOLUTION NMR | | Cite: | NMR solution structure of an oligodeoxynucleotide duplex containing the exocyclic lesion 3,N4-etheno-2'-deoxycytidine opposite thymidine: comparison with the duplex containing deoxyadenosine opposite the adduct.
Biochemistry, 35, 1996
|
|
1AW4
 
 | |
1AXV
 
 | | SOLUTION NMR STRUCTURE OF THE [BP]DA ADDUCT OPPOSITE DT IN A DNA DUPLEX, 6 STRUCTURES | | Descriptor: | 1,2,3-TRIHYDROXY-1,2,3,4-TETRAHYDROBENZO[A]PYRENE, DNA DUPLEX D(CTCTC-[BP]A-CTTCC)D(GGAAGTGAGAG) | | Authors: | Mao, B, Gu, Z, Gorin, A.A, Chen, J, Hingerty, B.E, Amid, S, Broyde, S, Geacintov, N.E, Patel, D.J. | | Deposit date: | 1997-10-21 | | Release date: | 1998-07-01 | | Last modified: | 2024-05-22 | | Method: | SOLUTION NMR | | Cite: | Solution structure of the (+)-cis-anti-benzo[a]pyrene-dA ([BP]dA) adduct opposite dT in a DNA duplex.
Biochemistry, 38, 1999
|
|
1AX6
 
 | | SOLUTION STRUCTURE OF THE [AF]-C8-DG ADDUCT OPPOSITE A-2 DELETION SITE IN THE NARI HOT SPOT SEQUENCE CONTEXT; NMR, 6 STRUCTURES | | Descriptor: | 2-AMINOFLUORENE, DNA DUPLEX D(CTCGGC-[AF]G-CCATC)D(GATGGCCGAG) | | Authors: | Mao, B, Gorin, A.A, Gu, Z, Hingerty, B.E, Broyde, S, Patel, D.J. | | Deposit date: | 1997-10-30 | | Release date: | 1998-07-01 | | Last modified: | 2024-05-22 | | Method: | SOLUTION NMR | | Cite: | Solution structure of the aminofluorene-intercalated conformer of the syn [AF]-C8-dG adduct opposite a--2 deletion site in the NarI hot spot sequence context.
Biochemistry, 36, 1997
|
|
1B6Y
 
 | | 3,N4-ETHENO-2'-DEOXYCYTIDINE OPPOSITE ADENINE IN AN 11-MER DUPLEX, SOLUTION STRUCTURE FROM NMR AND MOLECULAR DYNAMICS, 2 STRUCTURES | | Descriptor: | 5'-D(*CP*GP*TP*AP*CP*(EDC)P*CP*AP*TP*GP*C)-3', 5'-D(*GP*CP*AP*TP*GP*AP*GP*TP*AP*CP*G)-3' | | Authors: | Korobka, A, Cullinan, D, Cosman, M, Grollman, A.P, Patel, D.J, Eisenberg, M, De Los Santos, C. | | Deposit date: | 1999-01-19 | | Release date: | 1999-01-27 | | Last modified: | 2024-04-10 | | Method: | SOLUTION NMR | | Cite: | Solution structure of an oligodeoxynucleotide duplex containing the exocyclic lesion 3,N4-etheno-2'-deoxycytidine opposite 2'-deoxyadenosine, determined by NMR spectroscopy and restrained molecular dynamics.
Biochemistry, 35, 1996
|
|
1BIV
 
 | | BOVINE IMMUNODEFICIENCY VIRUS TAT-TAR COMPLEX, NMR, 5 STRUCTURES | | Descriptor: | TAR RNA, TAT PEPTIDE | | Authors: | Ye, X, Kumar, R.A, Patel, D.J. | | Deposit date: | 1996-06-12 | | Release date: | 1996-12-23 | | Last modified: | 2024-04-10 | | Method: | SOLUTION NMR | | Cite: | Molecular recognition in the bovine immunodeficiency virus Tat peptide-TAR RNA complex.
Chem.Biol., 2, 1995
|
|
6EDC
 
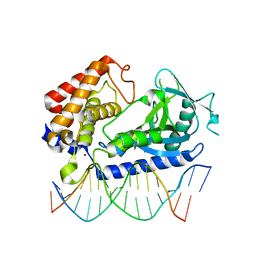 | | hcGAS-16bp dsDNA complex | | Descriptor: | Cyclic GMP-AMP synthase, DNA (5'-D(*AP*AP*AP*TP*TP*GP*CP*CP*GP*AP*AP*GP*AP*CP*GP*AP*A)-3'), DNA (5'-D(*TP*TP*CP*GP*TP*CP*TP*TP*CP*GP*GP*CP*AP*AP*T)-3'), ... | | Authors: | Xie, W, Lama, L, Adura, C, Glickman, J.F, Tuschl, T, Patel, D.J. | | Deposit date: | 2018-08-09 | | Release date: | 2019-05-29 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (2.712 Å) | | Cite: | Human cGAS catalytic domain has an additional DNA-binding interface that enhances enzymatic activity and liquid-phase condensation.
Proc.Natl.Acad.Sci.USA, 116, 2019
|
|
6EDB
 
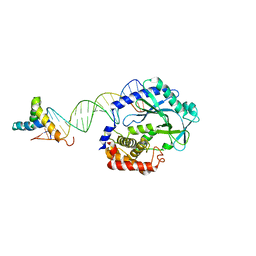 | | Crystal structure of SRY.hcGAS-21bp dsDNA complex | | Descriptor: | DNA (5'-D(*CP*GP*AP*TP*CP*CP*GP*GP*GP*AP*TP*CP*TP*AP*AP*AP*CP*AP*AP*TP*G)-3'), DNA (5'-D(*GP*CP*AP*TP*TP*GP*TP*TP*TP*AP*GP*AP*TP*CP*CP*CP*GP*GP*AP*TP*C)-3'), Sex-determining region Y protein,Cyclic GMP-AMP synthase, ... | | Authors: | Xie, W, Lama, L, Adura, C, Glickman, J.F, Tuschl, T, Patel, D.J. | | Deposit date: | 2018-08-09 | | Release date: | 2019-05-29 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (3.209 Å) | | Cite: | Human cGAS catalytic domain has an additional DNA-binding interface that enhances enzymatic activity and liquid-phase condensation.
Proc.Natl.Acad.Sci.USA, 116, 2019
|
|
1YVU
 
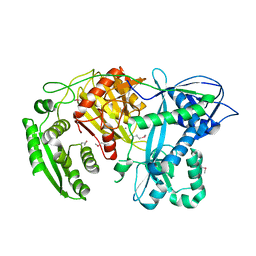 | | Crystal structure of A. aeolicus Argonaute | | Descriptor: | CALCIUM ION, hypothetical protein aq_1447 | | Authors: | Yuan, Y.R, Pei, Y, Ma, J.B, Kuryavyi, V, Zhadina, M, Meister, G, Chen, H.Y, Dauter, Z, Tuschl, T, Patel, D.J. | | Deposit date: | 2005-02-16 | | Release date: | 2005-08-09 | | Last modified: | 2024-11-20 | | Method: | X-RAY DIFFRACTION (2.9 Å) | | Cite: | Crystal structure of A. aeolicus Argonaute provides unique perspectives into the mechanism of guide strand-mediated mRNA cleavage
Mol.Cell, 19, 2005
|
|
2A5R
 
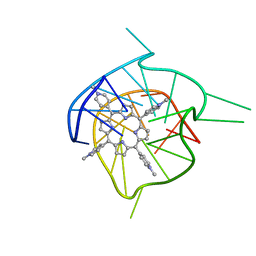 | | Complex of tetra-(4-n-methylpyridyl) porphin with monomeric parallel-stranded DNA tetraplex, snap-back 3+1 3' G-tetrad, single-residue chain reversal loops, GAG triad in the context of GAAG diagonal loop, C-MYC promoter, NMR, 6 struct. | | Descriptor: | (1Z,4Z,9Z,15Z)-5,10,15,20-tetrakis(1-methylpyridin-1-ium-4-yl)-21,23-dihydroporphyrin, 5'-D(*TP*GP*AP*GP*GP*GP*TP*GP*GP*IP*GP*AP*GP*GP*GP*TP*GP*GP*GP*GP*AP*AP*GP*G)-3' | | Authors: | Phan, A.T, Kuryavyi, V.V, Gaw, H.Y, Patel, D.J. | | Deposit date: | 2005-06-30 | | Release date: | 2005-07-26 | | Last modified: | 2024-05-01 | | Method: | SOLUTION NMR | | Cite: | Small-molecule interaction with a five-guanine-tract G-quadruplex structure from the human MYC promoter.
Nat.Chem.Biol., 1, 2005
|
|
4HRG
 
 | |
2A5P
 
 | | Monomeric parallel-stranded DNA tetraplex with snap-back 3+1 3' G-tetrad, single-residue chain reversal loops, GAG triad in the context of GAAG diagonal loop, NMR, 8 struct. | | Descriptor: | 5'-D(*TP*GP*AP*GP*GP*GP*TP*GP*GP*IP*GP*AP*GP*GP*GP*TP*GP*GP*GP*GP*AP*AP*GP*G)-3' | | Authors: | Phan, A.T, Kuryavyi, V.V, Gaw, H.Y, Patel, D.J. | | Deposit date: | 2005-06-30 | | Release date: | 2005-07-26 | | Last modified: | 2024-05-22 | | Method: | SOLUTION NMR | | Cite: | Small-molecule interaction with a five-guanine-tract G-quadruplex structure from the human MYC promoter.
Nat.Chem.Biol., 1, 2005
|
|
2AQY
 
 | |
2ASD
 
 | | oxoG-modified Insertion Ternary Complex | | Descriptor: | 2'-DEOXYCYTIDINE-5'-TRIPHOSPHATE, 5'-D(*CP*T*AP*AP*CP*(8OG)P*CP*TP*AP*CP*CP*AP*TP*CP*CP*AP*AP*CP*C)-3', 5'-D(*GP*GP*TP*TP*GP*GP*AP*TP*GP*GP*TP*AP*(DDG))-3', ... | | Authors: | Rechkoblit, O, Malinina, L, Cheng, Y, Kuryavyi, V, Broyde, S, Geacintov, N.E, Patel, D.J. | | Deposit date: | 2005-08-23 | | Release date: | 2006-01-10 | | Last modified: | 2023-08-23 | | Method: | X-RAY DIFFRACTION (1.95 Å) | | Cite: | Stepwise Translocation of Dpo4 Polymerase during Error-Free Bypass of an oxoG Lesion
Plos Biol., 4, 2006
|
|
2AU0
 
 | | Unmodified preinsertion binary complex | | Descriptor: | 5'-D(*CP*TP*AP*AP*CP*G*CP*TP*AP*CP*CP*AP*TP*CP*CP*AP*AP*CP*C)-3', 5'-D(*GP*GP*TP*TP*GP*GP*AP*TP*GP*GP*TP*AP*(DDG))-3', CALCIUM ION, ... | | Authors: | Rechkoblit, O, Malinina, L, Cheng, Y, Kuryavyi, V, Broyde, S, Geacintov, N.E, Patel, D.J. | | Deposit date: | 2005-08-26 | | Release date: | 2006-01-10 | | Last modified: | 2023-08-23 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | Stepwise Translocation of Dpo4 Polymerase during Error-Free Bypass of an oxoG Lesion
Plos Biol., 4, 2006
|
|
1YKQ
 
 | | Crystal structure of Diels-Alder ribozyme | | Descriptor: | CADMIUM ION, Diels-Alder ribozyme, MAGNESIUM ION | | Authors: | Serganov, A, Keiper, S, Malinina, L, Tereshko, V, Skripkin, E, Hobartner, C, Polonskaia, A, Phan, A.T, Wombacher, R, Micura, R, Dauter, Z, Jaschke, A, Patel, D.J. | | Deposit date: | 2005-01-18 | | Release date: | 2005-02-22 | | Last modified: | 2023-08-23 | | Method: | X-RAY DIFFRACTION (3.5 Å) | | Cite: | Structural basis for Diels-Alder ribozyme-catalyzed carbon-carbon bond formation.
Nat.Struct.Mol.Biol., 12, 2005
|
|
1YKV
 
 | | Crystal structure of the Diels-Alder ribozyme complexed with the product of the reaction between N-pentylmaleimide and covalently attached 9-hydroxymethylanthracene | | Descriptor: | (3AS,9AS)-2-PENTYL-4-HYDROXYMETHYL-3A,4,9,9A-TETRAHYDRO-4,9[1',2']-BENZENO-1H-BENZ[F]ISOINDOLE-1,3(2H)-DIONE, Diels-Alder ribozyme, MAGNESIUM ION | | Authors: | Serganov, A, Keiper, S, Malinina, L, Tereshko, V, Skripkin, E, Hobartner, C, Polonskaia, A, Phan, A.T, Wombacher, R, Micura, R, Dauter, Z, Jaschke, A, Patel, D.J. | | Deposit date: | 2005-01-18 | | Release date: | 2005-02-22 | | Last modified: | 2023-08-23 | | Method: | X-RAY DIFFRACTION (3.3 Å) | | Cite: | Structural basis for Diels-Alder ribozyme-catalyzed carbon-carbon bond formation.
Nat.Struct.Mol.Biol., 12, 2005
|
|
1YTY
 
 | | Structural basis for recognition of UUUOH 3'-terminii of nascent RNA pol III transcripts by La autoantigen | | Descriptor: | 5'-R(*UP*GP*CP*UP*GP*UP*UP*UP*U)-3', Lupus La protein | | Authors: | Teplova, M, Yuan, Y.R, Ilin, S, Malinina, L, Phan, A.T, Teplov, A, Patel, D.J. | | Deposit date: | 2005-02-11 | | Release date: | 2006-01-17 | | Last modified: | 2024-11-20 | | Method: | X-RAY DIFFRACTION (2.29 Å) | | Cite: | Structural Basis for Recognition and Sequestration of UUU(OH) 3' Temini of Nascent RNA Polymerase III Transcripts by La, a Rheumatic Disease Autoantigen.
Mol.Cell, 21, 2006
|
|
4JVH
 
 | | Structure of the star domain of quaking protein in complex with RNA | | Descriptor: | Protein quaking, RNA (5'-R(*UP*UP*CP*AP*CP*UP*AP*AP*CP*AP*A)-3'), SULFATE ION | | Authors: | Teplova, M, Hafner, M, Teplov, D, Essig, K, Tuschl, T, Patel, D.J. | | Deposit date: | 2013-03-25 | | Release date: | 2013-05-08 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (3.501 Å) | | Cite: | Structure-function studies of STAR family Quaking proteins bound to their in vivo RNA target sites.
Genes Dev., 27, 2013
|
|
207D
 
 | | SOLUTION STRUCTURE OF MITHRAMYCIN DIMERS BOUND TO PARTIALLY OVERLAPPING SITES ON DNA | | Descriptor: | 1,2-HYDRO-1-OXY-3,4-HYDRO-3-(1-METHOXY-2-OXY-3,4-DIHYDROXYPENTYL)-8,9-DIHYROXY-7-METHYLANTHRACENE, 2,6-dideoxy-3-C-methyl-beta-D-ribo-hexopyranose-(1-3)-2,6-dideoxy-beta-D-galactopyranose-(1-3)-beta-D-Olivopyranose, DNA (5'-D(*TP*AP*GP*CP*TP*AP*GP*CP*TP*A)-3'), ... | | Authors: | Sastry, M, Fiala, R, Patel, D.J. | | Deposit date: | 1995-04-20 | | Release date: | 1995-09-15 | | Last modified: | 2024-05-01 | | Method: | SOLUTION NMR | | Cite: | Solution structure of mithramycin dimers bound to partially overlapping sites on DNA.
J.Mol.Biol., 251, 1995
|
|
4JOL
 
 | |
