6FAI
 
 | | Structure of a eukaryotic cytoplasmic pre-40S ribosomal subunit | | Descriptor: | 20S ribosomal RNA, 40S ribosomal protein S0-A, 40S ribosomal protein S1-A, ... | | Authors: | Scaiola, A, Pena, C, Weisser, M, Boehringer, D, Leibundgut, M, Klingauf-Nerurkar, P, Gerhardy, S, Panse, V.G, Ban, N. | | Deposit date: | 2017-12-15 | | Release date: | 2018-02-28 | | Last modified: | 2024-05-15 | | Method: | ELECTRON MICROSCOPY (3.4 Å) | | Cite: | Structure of a eukaryotic cytoplasmic pre-40S ribosomal subunit.
EMBO J., 37, 2018
|
|
7ASD
 
 | | Structure of native royal jelly filaments | | Descriptor: | (3beta,14beta,17alpha)-ergosta-5,24(28)-dien-3-ol, 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, ... | | Authors: | Mattei, S, Ban, A, Picenoni, A, Leibundgut, M, Glockshuber, R, Boehringer, D. | | Deposit date: | 2020-10-27 | | Release date: | 2020-12-30 | | Last modified: | 2024-11-13 | | Method: | ELECTRON MICROSCOPY (3.5 Å) | | Cite: | Structure of native glycolipoprotein filaments in honeybee royal jelly.
Nat Commun, 11, 2020
|
|
6ZOK
 
 | | SARS-CoV-2-Nsp1-40S complex, focused on body | | Descriptor: | 18S ribosomal RNA, 40S ribosomal protein S11, 40S ribosomal protein S13, ... | | Authors: | Schubert, K, Karousis, E.D, Jomaa, A, Scaiola, A, Echeverria, B, Gurzeler, L.-A, Leibundgut, M, Thiel, V, Muehlemann, O, Ban, N. | | Deposit date: | 2020-07-07 | | Release date: | 2020-07-29 | | Last modified: | 2024-11-13 | | Method: | ELECTRON MICROSCOPY (2.8 Å) | | Cite: | SARS-CoV-2 Nsp1 binds the ribosomal mRNA channel to inhibit translation.
Nat.Struct.Mol.Biol., 27, 2020
|
|
5NCO
 
 | | Quaternary complex between SRP, SR, and SecYEG bound to the translating ribosome | | Descriptor: | 23S rRNA, 4.5S SRP RNA (Ffs), 50S ribosomal protein L10, ... | | Authors: | Jomaa, A, Hwang Fu, Y, Boerhinger, D, Leibundgut, M, Shan, S.O, Ban, N. | | Deposit date: | 2017-03-06 | | Release date: | 2017-05-24 | | Last modified: | 2018-03-28 | | Method: | ELECTRON MICROSCOPY (4.8 Å) | | Cite: | Structure of the quaternary complex between SRP, SR, and translocon bound to the translating ribosome.
Nat Commun, 8, 2017
|
|
7AVE
 
 | | Perdeuterated refolded hen egg-white lysozyme at 100 K | | Descriptor: | ACETATE ION, Lysozyme C, NITRATE ION | | Authors: | Ramos, J, Laux, V, Haertlein, M, Erba Boeri, E, Forsyth, V.T, Larsen, S, Mossou, E, Langkilde, A.E. | | Deposit date: | 2020-11-05 | | Release date: | 2021-05-12 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (0.98 Å) | | Cite: | Structural insights into protein folding, stability and activity using in vivo perdeuteration of hen egg-white lysozyme.
Iucrj, 8, 2021
|
|
7AVF
 
 | | Triclinic hydrogenated hen egg-white lysozyme at 100 K (control) | | Descriptor: | ACETATE ION, Lysozyme, NITRATE ION | | Authors: | Ramos, J, Laux, V, Haertlein, M, Erba Boeri, E, Forsyth, V.T, Mossou, E, Larsen, S, Langkilde, A.E. | | Deposit date: | 2020-11-05 | | Release date: | 2021-05-12 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (1 Å) | | Cite: | Structural insights into protein folding, stability and activity using in vivo perdeuteration of hen egg-white lysozyme.
Iucrj, 8, 2021
|
|
7AVG
 
 | | Perdeuterated hen egg-white lysozyme at 100 K | | Descriptor: | ACETATE ION, Lysozyme, NITRATE ION | | Authors: | Ramos, J, Laux, V, Haertlein, M, Erba Boeri, E, Forsyth, V.T, Mossou, E, Larsen, S, Langkilde, A.E. | | Deposit date: | 2020-11-05 | | Release date: | 2021-05-12 | | Last modified: | 2024-11-13 | | Method: | X-RAY DIFFRACTION (1 Å) | | Cite: | Structural insights into protein folding, stability and activity using in vivo perdeuteration of hen egg-white lysozyme.
Iucrj, 8, 2021
|
|
6E57
 
 | | Bacteroides ovatus mixed-linkage glucan utilization locus (MLGUL) SGBP-B in complex with mixed-linkage heptasaccharide | | Descriptor: | 1,2-ETHANEDIOL, PHOSPHATE ION, SODIUM ION, ... | | Authors: | Koropatkin, N.M, Schnizlein, M, Bahr, C.M.E. | | Deposit date: | 2018-07-19 | | Release date: | 2019-05-29 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (2.71 Å) | | Cite: | Surface glycan-binding proteins are essential for cereal beta-glucan utilization by the human gut symbiont Bacteroides ovatus.
Cell.Mol.Life Sci., 76, 2019
|
|
6GB2
 
 | | Unique features of mammalian mitochondrial translation initiation revealed by cryo-EM. This file contains the 39S ribosomal subunit. | | Descriptor: | 'Mitochondrial ribosomal protein L30, 'Mitochondrial ribosomal protein L55, 'Mitochondrial ribosomal protein L59, ... | | Authors: | Kummer, E, Leibundgut, M, Boehringer, D, Ban, N. | | Deposit date: | 2018-04-13 | | Release date: | 2018-08-08 | | Last modified: | 2024-05-15 | | Method: | ELECTRON MICROSCOPY (3.2 Å) | | Cite: | Unique features of mammalian mitochondrial translation initiation revealed by cryo-EM.
Nature, 560, 2018
|
|
6GAZ
 
 | | Unique features of mammalian mitochondrial translation initiation revealed by cryo-EM. This file contains the 28S ribosomal subunit. | | Descriptor: | 12S ribosomal RNA, mitochondrial, 28S ribosomal protein S18b, ... | | Authors: | Kummer, E, Leibundgut, M, Boehringer, D, Ban, N. | | Deposit date: | 2018-04-13 | | Release date: | 2018-08-08 | | Last modified: | 2024-11-13 | | Method: | ELECTRON MICROSCOPY (3.1 Å) | | Cite: | Unique features of mammalian mitochondrial translation initiation revealed by cryo-EM.
Nature, 560, 2018
|
|
6GAW
 
 | | Unique features of mammalian mitochondrial translation initiation revealed by cryo-EM. This file contains the complete 55S ribosome. | | Descriptor: | 12S ribosomal RNA, mitochondrial, 16S ribosomal RNA, ... | | Authors: | Kummer, E, Leibundgut, M, Boehringer, D, Ban, N. | | Deposit date: | 2018-04-13 | | Release date: | 2018-08-22 | | Last modified: | 2019-12-11 | | Method: | ELECTRON MICROSCOPY (3.2 Å) | | Cite: | Unique features of mammalian mitochondrial translation initiation revealed by cryo-EM.
Nature, 560, 2018
|
|
6BPH
 
 | | Crystal structure of the chromodomain of RBBP1 | | Descriptor: | AT-rich interactive domain-containing protein 4A, UNKNOWN ATOM OR ION | | Authors: | Liu, Y, Tempel, W, Bountra, C, Arrowsmith, C.H, Edwards, A.M, Min, J, Structural Genomics Consortium (SGC) | | Deposit date: | 2017-11-23 | | Release date: | 2017-12-20 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (1.85 Å) | | Cite: | Crystal structure of chromo barrel domain of RBBP1.
Biochem. Biophys. Res. Commun., 496, 2018
|
|
6C1Y
 
 | | mbd of human mecp2 in complex with methylated DNA | | Descriptor: | 12-mer DNA, Methyl-CpG-binding protein 2, UNKNOWN ATOM OR ION | | Authors: | Liu, K, Bian, C, Tempel, W, Wernimont, A.K, Arrowsmith, C.H, Bountra, C, Edwards, A.M, Min, J, Structural Genomics Consortium (SGC) | | Deposit date: | 2018-01-05 | | Release date: | 2018-02-14 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Plasticity at the DNA recognition site of the MeCP2 mCG-binding domain.
Biochim Biophys Acta Gene Regul Mech, 1862, 2019
|
|
9FQ0
 
 | | Human NatA-NAC-MAP1 80S ribosome complex | | Descriptor: | 28S rRNA, 5.8S rRNA, 60S ribosomal protein L19, ... | | Authors: | Klein, M.A, Wild, K, Sinning, I. | | Deposit date: | 2024-06-14 | | Release date: | 2024-07-03 | | Last modified: | 2024-10-02 | | Method: | ELECTRON MICROSCOPY (4.67 Å) | | Cite: | Multi-protein assemblies orchestrate co-translational enzymatic processing on the human ribosome.
Nat Commun, 15, 2024
|
|
9FPZ
 
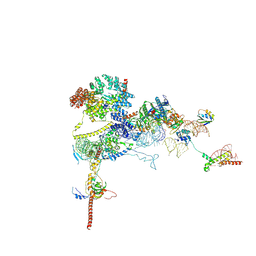 | | Human NatA-MAP2 80S ribosome complex | | Descriptor: | 28S rRNA, 5.8S rRNA (58-MER), 60S ribosomal protein L19, ... | | Authors: | Klein, M.A, Wild, K, Sinning, I. | | Deposit date: | 2024-06-14 | | Release date: | 2024-07-03 | | Last modified: | 2024-10-02 | | Method: | ELECTRON MICROSCOPY (2.69 Å) | | Cite: | Multi-protein assemblies orchestrate co-translational enzymatic processing on the human ribosome.
Nat Commun, 15, 2024
|
|
5F9E
 
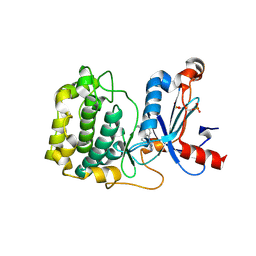 | | Structure of Protein Kinase C theta with compound 10: 2,2-dimethyl-7-(2-oxidanylidene-3~{H}-imidazo[4,5-b]pyridin-1-yl)-1-(phenylmethyl)-3~{H}-quinazolin-4-one | | Descriptor: | 2,2-dimethyl-7-(2-oxidanylidene-3~{H}-imidazo[4,5-b]pyridin-1-yl)-1-(phenylmethyl)-3~{H}-quinazolin-4-one, Protein kinase C theta type | | Authors: | Klein, M. | | Deposit date: | 2015-12-09 | | Release date: | 2016-05-11 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Discovery and optimization of 1,7-disubstituted-2,2-dimethyl-2,3-dihydroquinazolin-4(1H)-ones as potent and selective PKC theta inhibitors.
Bioorg.Med.Chem., 24, 2016
|
|
5VFC
 
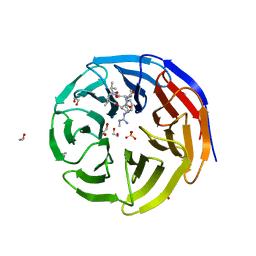 | | WDR5 bound to inhibitor MM-589 | | Descriptor: | 1,2-ETHANEDIOL, N-{(3R,6S,9S,12R)-6-ethyl-12-methyl-9-[3-(N'-methylcarbamimidamido)propyl]-2,5,8,11-tetraoxo-3-phenyl-1,4,7,10-tetraazacyclotetradecan-12-yl}-2-methylpropanamide, SULFATE ION, ... | | Authors: | Stuckey, J.A. | | Deposit date: | 2017-04-07 | | Release date: | 2017-06-28 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (1.64 Å) | | Cite: | Discovery of a Highly Potent, Cell-Permeable Macrocyclic Peptidomimetic (MM-589) Targeting the WD Repeat Domain 5 Protein (WDR5)-Mixed Lineage Leukemia (MLL) Protein-Protein Interaction.
J. Med. Chem., 60, 2017
|
|
3D0Z
 
 | |
6CNP
 
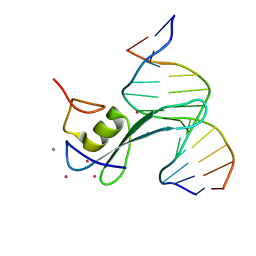 | |
6KIX
 
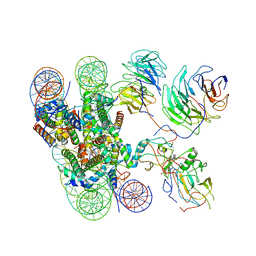 | | Cryo-EM structure of human MLL1-NCP complex, binding mode1 | | Descriptor: | DNA (145-MER), GLUTAMINE, Histone H2A, ... | | Authors: | Huang, J, Xue, H, Yao, T. | | Deposit date: | 2019-07-20 | | Release date: | 2019-09-11 | | Last modified: | 2024-03-27 | | Method: | ELECTRON MICROSCOPY (4.1 Å) | | Cite: | Structural basis of nucleosome recognition and modification by MLL methyltransferases.
Nature, 573, 2019
|
|
6KIV
 
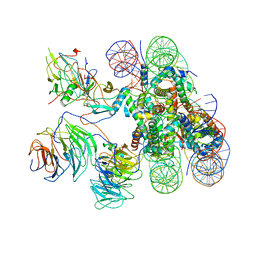 | | Cryo-EM structure of human MLL1-ubNCP complex (4.0 angstrom) | | Descriptor: | DNA (145-MER), Histone H2A, Histone H2B 1.1, ... | | Authors: | Huang, J, Xue, H, Yao, T. | | Deposit date: | 2019-07-20 | | Release date: | 2019-09-11 | | Last modified: | 2024-03-27 | | Method: | ELECTRON MICROSCOPY (4 Å) | | Cite: | Structural basis of nucleosome recognition and modification by MLL methyltransferases.
Nature, 573, 2019
|
|
6KIU
 
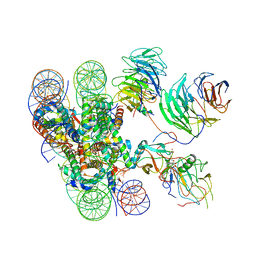 | | Cryo-EM structure of human MLL1-ubNCP complex (3.2 angstrom) | | Descriptor: | DNA (145-MER), GLUTAMINE, Histone H2A, ... | | Authors: | Huang, J, Xue, H, Yao, T. | | Deposit date: | 2019-07-20 | | Release date: | 2019-09-11 | | Last modified: | 2024-03-27 | | Method: | ELECTRON MICROSCOPY (3.2 Å) | | Cite: | Structural basis of nucleosome recognition and modification by MLL methyltransferases.
Nature, 573, 2019
|
|
6KIZ
 
 | | Cryo-EM structure of human MLL1-NCP complex, binding mode2 | | Descriptor: | DNA (145-MER), Histone H2A, Histone H2B 1.1, ... | | Authors: | Huang, J, Xue, H, Yao, T. | | Deposit date: | 2019-07-20 | | Release date: | 2019-09-11 | | Last modified: | 2024-03-27 | | Method: | ELECTRON MICROSCOPY (4.5 Å) | | Cite: | Structural basis of nucleosome recognition and modification by MLL methyltransferases.
Nature, 573, 2019
|
|
6KIW
 
 | | Cryo-EM structure of human MLL3-ubNCP complex (4.0 angstrom) | | Descriptor: | DNA (144-MER), DNA (145-MER), Histone H2A, ... | | Authors: | Huang, J, Xue, H, Yao, T. | | Deposit date: | 2019-07-20 | | Release date: | 2019-09-11 | | Last modified: | 2024-03-27 | | Method: | ELECTRON MICROSCOPY (4 Å) | | Cite: | Structural basis of nucleosome recognition and modification by MLL methyltransferases.
Nature, 573, 2019
|
|
6XCG
 
 | | Histone-lysine N-methyltransferase NSD2-PWWP1 with compound UNC6934 | | Descriptor: | Histone-lysine N-methyltransferase NSD2, N-cyclopropyl-3-oxo-N-({4-[(pyrimidin-4-yl)carbamoyl]phenyl}methyl)-3,4-dihydro-2H-1,4-benzoxazine-7-carboxamide, UNKNOWN ATOM OR ION | | Authors: | Zhou, M.Q, Dong, A, Ingerman, L.A, Hanley, R.P, Bountra, C, Arrowsmith, C.H, Edwards, A.M, Min, J, Structural Genomics Consortium (SGC) | | Deposit date: | 2020-06-08 | | Release date: | 2020-07-22 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (1.64 Å) | | Cite: | A chemical probe targeting the PWWP domain alters NSD2 nucleolar localization.
Nat.Chem.Biol., 18, 2022
|
|
