2VNF
 
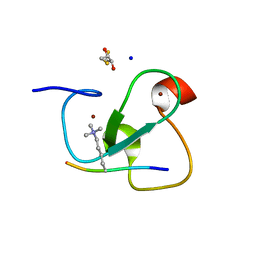 | | MOLECULAR BASIS OF HISTONE H3K4ME3 RECOGNITION BY ING4 | | Descriptor: | (2R,3S)-1,4-DIMERCAPTOBUTANE-2,3-DIOL, 2,3-DIHYDROXY-1,4-DITHIOBUTANE, HISTONE H3, ... | | Authors: | Palacios, A, Munoz, I.G, Pantoja-Uceda, D, Marcaida, M.J, Torres, D, Martin-Garcia, J.M, Luque, I, Montoya, G, Blanco, F.J. | | Deposit date: | 2008-02-04 | | Release date: | 2008-04-01 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (1.76 Å) | | Cite: | Molecular Basis of Histone H3K4Me3 Recognition by Ing4
J.Biol.Chem., 283, 2008
|
|
6PBV
 
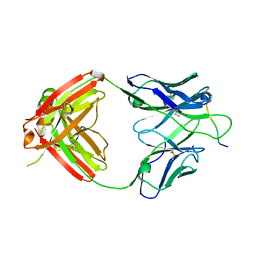 | | Crystal structure of Fab668 complex | | Descriptor: | 1,2-ETHANEDIOL, Fab668 heavy chain, Fab668 light chain, ... | | Authors: | Oyen, D, Wilson, I.A. | | Deposit date: | 2019-06-14 | | Release date: | 2020-03-04 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (1.566 Å) | | Cite: | Structure and mechanism of monoclonal antibody binding to the junctional epitope of Plasmodium falciparum circumsporozoite protein.
Plos Pathog., 16, 2020
|
|
8ROR
 
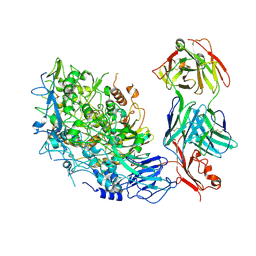 | | Single-particle cryo-EM of Mycoplasma pneumoniae adhesin P1 complexed with the anti-adhesive Fab fragment. | | Descriptor: | Adhesin P1, Heavy Chain Fab, Light Chain Fab | | Authors: | Vizarraga, D, Kawamoto, A, Marcos-Silva, M, Fita, I, Miyata, M, Pinyol, J, Namba, K, Kenri, T. | | Deposit date: | 2024-01-12 | | Release date: | 2025-03-05 | | Last modified: | 2025-07-09 | | Method: | ELECTRON MICROSCOPY (2.39 Å) | | Cite: | Dynamics of the adhesion complex of the human pathogens Mycoplasma pneumoniae and Mycoplasma genitalium.
Plos Pathog., 21, 2025
|
|
2UUH
 
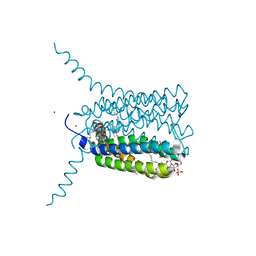 | | Crystal structure of Human Leukotriene C4 Synthase in complex with substrate glutathione | | Descriptor: | DODECYL-ALPHA-D-MALTOSIDE, GLUTATHIONE, LEUKOTRIENE C4 SYNTHASE, ... | | Authors: | Martinez Molina, D, Wetterholm, A, Kohl, A, McCarthy, A.A, Niegowski, D, Ohlson, E, Hammarberg, T, Eshaghi, S, Haeggstrom, J.Z, Nordlund, P. | | Deposit date: | 2007-03-02 | | Release date: | 2007-07-17 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (2.15 Å) | | Cite: | Structural Basis for Synthesis of Inflammatory Mediators by Human Leukotriene C4 Synthase.
Nature, 448, 2007
|
|
2VT7
 
 | | Native Torpedo californica acetylcholinesterase collected with a cumulated dose of 800000 Gy | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, ACETYLCHOLINESTERASE, CHLORIDE ION, ... | | Authors: | Colletier, J.P, Bourgeois, D, Sanson, B, Fournier, D, Sussman, J.L, Silman, I, Weik, M. | | Deposit date: | 2008-05-09 | | Release date: | 2008-07-22 | | Last modified: | 2024-11-13 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Shoot-and-Trap: Use of Specific X-Ray Damage to Study Structural Protein Dynamics by Temperature-Controlled Cryo-Crystallography.
Proc.Natl.Acad.Sci.USA, 105, 2008
|
|
6PLF
 
 | | Crystal structure of human PHGDH complexed with Compound 1 | | Descriptor: | 1,2-ETHANEDIOL, 4-{(1S)-1-[(5-chloro-6-{[(5S)-2-oxo-1,3-oxazolidin-5-yl]methoxy}-1H-indole-2-carbonyl)amino]-2-hydroxyethyl}benzoic acid, D-3-phosphoglycerate dehydrogenase | | Authors: | Olland, A, Lakshminarasimhan, D, White, A, Suto, R.K. | | Deposit date: | 2019-06-30 | | Release date: | 2019-07-24 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Inhibition of 3-phosphoglycerate dehydrogenase (PHGDH) by indole amides abrogates de novo serine synthesis in cancer cells.
Bioorg.Med.Chem.Lett., 29, 2019
|
|
7NRU
 
 | |
2VY7
 
 | | The 627-domain from influenza A virus polymerase PB2 subunit | | Descriptor: | POLYMERASE BASIC PROTEIN 2 | | Authors: | Tarendeau, F, Crepin, T, Guilligay, D, Ruigrok, R, Cusack, S, Hart, D. | | Deposit date: | 2008-07-18 | | Release date: | 2008-09-09 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (1.53 Å) | | Cite: | Host Determinant Residue Lysine 627 Lies on the Surface of a Discrete, Folded Domain of Influenza Virus Polymerase Pb2 Subunit
Plos Pathog., 4, 2008
|
|
8V3U
 
 | | ACAD11 D220A with 4-hydroxyvaleryl-CoA | | Descriptor: | Acyl-Coenzyme A dehydrogenase family, member 11, FLAVIN-ADENINE DINUCLEOTIDE | | Authors: | Zhang, J, Bartlett, A, Rashan, E, Yuan, P, Pagliarini, D. | | Deposit date: | 2023-11-28 | | Release date: | 2024-12-11 | | Last modified: | 2025-07-02 | | Method: | ELECTRON MICROSCOPY (2.6 Å) | | Cite: | ACAD10 and ACAD11 enable mammalian 4-hydroxy acid lipid catabolism.
Nat.Struct.Mol.Biol., 2025
|
|
8V3V
 
 | | ACAD11 D753N with 4-phosphovaleryl-CoA | | Descriptor: | Acyl-Coenzyme A dehydrogenase family, member 11, FLAVIN-ADENINE DINUCLEOTIDE | | Authors: | Zhang, J, Bartlett, A, Rashan, E, Yuan, P, Pagliarini, D. | | Deposit date: | 2023-11-28 | | Release date: | 2024-12-11 | | Last modified: | 2025-07-02 | | Method: | ELECTRON MICROSCOPY (3.6 Å) | | Cite: | ACAD10 and ACAD11 enable mammalian 4-hydroxy acid lipid catabolism.
Nat.Struct.Mol.Biol., 2025
|
|
8C7Z
 
 | | Crystal structure of the ACVR1 (ALK2) kinase in complex with the compound M4K2308 | | Descriptor: | 1,2-ETHANEDIOL, 9-piperazin-1-yl-4-(3,4,5-trimethoxyphenyl)-5,6-dihydro-[1]benzoxepino[5,4-c]pyridine, AMMONIUM ION, ... | | Authors: | Cros, J, Williams, E.P, Sweeney, M.N, Smil, D, Gonzalez-Alvarez, H, Al-awar, R, Bullock, A.N. | | Deposit date: | 2023-01-18 | | Release date: | 2023-02-01 | | Last modified: | 2024-08-14 | | Method: | X-RAY DIFFRACTION (2.23 Å) | | Cite: | Discovery of Conformationally Constrained ALK2 Inhibitors.
J.Med.Chem., 67, 2024
|
|
8C7W
 
 | | Crystal structure of the ACVR1 (ALK2) kinase in complex with the compound M4K2304 | | Descriptor: | 1,2-ETHANEDIOL, 6-methyl-9-piperazin-1-yl-4-(3,4,5-trimethoxyphenyl)-5,7-dihydropyrido[4,3-d][2]benzazepine, Activin receptor type I, ... | | Authors: | Cros, J, Williams, E.P, Sweeney, M.N, Smil, D, Gonzalez-Alvarez, H, Al-awar, R, Bullock, A.N. | | Deposit date: | 2023-01-17 | | Release date: | 2023-02-08 | | Last modified: | 2024-08-14 | | Method: | X-RAY DIFFRACTION (2.26 Å) | | Cite: | Discovery of Conformationally Constrained ALK2 Inhibitors.
J.Med.Chem., 67, 2024
|
|
7SGT
 
 | |
2W0S
 
 | | Crystal structure of vaccinia virus thymidylate kinase bound to brivudin-5'-monophosphate | | Descriptor: | (E)-5-(2-BROMOVINYL)-2'-DEOXYURIDINE-5'-MONOPHOSPHATE, MAGNESIUM ION, SULFATE ION, ... | | Authors: | Caillat, C, Topalis, D, Agrofoglio, L.A, Pochet, S, Balzarini, J, Deville-Bonne, D, Meyer, P. | | Deposit date: | 2008-10-08 | | Release date: | 2008-10-21 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (2.918 Å) | | Cite: | Crystal Structure of Poxvirus Thymidylate Kinase: An Unexpected Dimerization Has Implications for Antiviral Therapy
Proc.Natl.Acad.Sci.USA, 105, 2008
|
|
2VY8
 
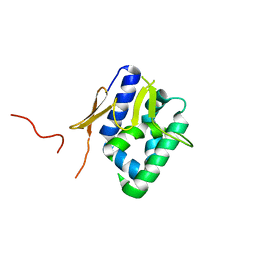 | | The 627-domain from influenza A virus polymerase PB2 subunit with Glu- 627 | | Descriptor: | POLYMERASE BASIC PROTEIN 2 | | Authors: | Tarendeau, F, Crepin, T, Guilligay, D, Ruigrok, R, Cusack, S, Hart, D. | | Deposit date: | 2008-07-19 | | Release date: | 2008-09-09 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (1.2 Å) | | Cite: | Host Determinant Residue Lysine 627 Lies on the Surface of a Discrete, Folded Domain of Influenza Virus Polymerase Pb2 Subunit
Plos Pathog., 4, 2008
|
|
3ENC
 
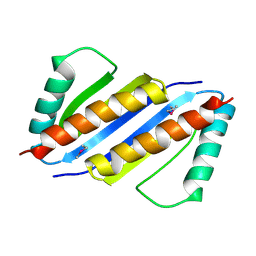 | | Crystal structure of Pyrococcus furiosus PCC1 dimer | | Descriptor: | protein PCC1 | | Authors: | Neculai, D. | | Deposit date: | 2008-09-25 | | Release date: | 2008-10-28 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (2.6304 Å) | | Cite: | Atomic structure of the KEOPS complex: an ancient protein kinase-containing molecular machine.
Mol.Cell, 32, 2008
|
|
7W8M
 
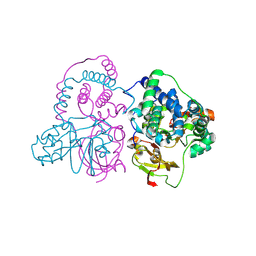 | | Crystal structure of Co-type nitrile hydratase mutant from Pseudomonas thermophila - A129R | | Descriptor: | COBALT (II) ION, Cobalt-containing nitrile hydratase subunit beta, Nitrile hydratase | | Authors: | Ma, D, Cheng, Z.Y, Hou, X.D, Peplowski, L, Lai, Q.P, Fu, K, Yin, D.J, Rao, Y.J, Zhou, Z.M. | | Deposit date: | 2021-12-08 | | Release date: | 2022-08-24 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Insight into the broadened substrate scope of nitrile hydratase by static and dynamic structure analysis.
Chem Sci, 13, 2022
|
|
3EOV
 
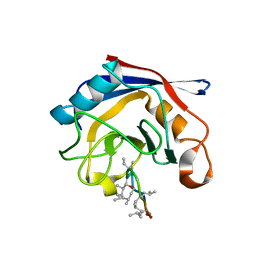 | | Crystal structure of cyclophilin from Leishmania donovani ligated with cyclosporin A | | Descriptor: | CYCLOSPORIN A, PEPTIDYL-PROLYL CIS-TRANS ISOMERASE | | Authors: | Venugopal, V, Dasgupta, D, Datta, A.K, Banerjee, R. | | Deposit date: | 2008-09-29 | | Release date: | 2008-11-11 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Structure of Cyclophilin from Leishmania Donovani Bound to Cyclosporin at 2.6 A Resolution: Correlation between Structure and Thermodynamic Data.
Acta Crystallogr.,Sect.D, 65, 2009
|
|
3ENO
 
 | |
7ABK
 
 | | Helical structure of PspA | | Descriptor: | Chloroplast membrane-associated 30 kD protein | | Authors: | Junglas, B, Huber, S.T, Mann, D, Heidler, T, Clarke, M, Schneider, D, Sachse, C. | | Deposit date: | 2020-09-07 | | Release date: | 2021-08-04 | | Last modified: | 2024-07-10 | | Method: | ELECTRON MICROSCOPY (3.6 Å) | | Cite: | PspA adopts an ESCRT-III-like fold and remodels bacterial membranes.
Cell, 184, 2021
|
|
7QCX
 
 | | Two-state liquid NMR Structure of a PDZ2 Domain from hPTP1E, apo form | | Descriptor: | Tyrosine-protein phosphatase non-receptor type 13 | | Authors: | Ashkinadze, D, Kadavath, H, Chi, C, Friedmann, M, Strotz, D, Kumari, P, Minges, M, Cadalbert, R, Koenigl, S, Guentert, P, Voegeli, B, Riek, R. | | Deposit date: | 2021-11-25 | | Release date: | 2022-09-07 | | Last modified: | 2024-06-19 | | Method: | SOLUTION NMR | | Cite: | Atomic resolution protein allostery from the multi-state structure of a PDZ domain.
Nat Commun, 13, 2022
|
|
7QCY
 
 | | Two-state liquid NMR Structure of a PDZ2 Domain from hPTP1E, complexed with RA-GEF2 peptide | | Descriptor: | Tyrosine-protein phosphatase non-receptor type 13 | | Authors: | Ashkinadze, D, Kadavath, H, Chi, C, Friedmann, M, Strotz, D, Kumari, P, Minges, M, Cadalbert, R, Koenigl, S, Guentert, P, Voegeli, B, Riek, R. | | Deposit date: | 2021-11-25 | | Release date: | 2022-09-07 | | Last modified: | 2024-06-19 | | Method: | SOLUTION NMR | | Cite: | Atomic resolution protein allostery from the multi-state structure of a PDZ domain.
Nat Commun, 13, 2022
|
|
6Q9C
 
 | | Crystal structure of Aquifex aeolicus NADH-quinone oxidoreductase subunits NuoE and NuoF bound to NADH under anaerobic conditions | | Descriptor: | 1,4-DIHYDRONICOTINAMIDE ADENINE DINUCLEOTIDE, FE2/S2 (INORGANIC) CLUSTER, FLAVIN MONONUCLEOTIDE, ... | | Authors: | Wohlwend, D, Gerhardt, S, Gnandt, E, Friedrich, T. | | Deposit date: | 2018-12-17 | | Release date: | 2019-06-26 | | Last modified: | 2024-05-15 | | Method: | X-RAY DIFFRACTION (1.78 Å) | | Cite: | A mechanism to prevent production of reactive oxygen species by Escherichia coli respiratory complex I.
Nat Commun, 10, 2019
|
|
6Q9G
 
 | | Crystal structure of reduced Aquifex aeolicus NADH-quinone oxidoreductase subunits NuoE G129D and NuoF bound to NADH | | Descriptor: | 1,4-DIHYDRONICOTINAMIDE ADENINE DINUCLEOTIDE, FE2/S2 (INORGANIC) CLUSTER, FLAVIN MONONUCLEOTIDE, ... | | Authors: | Wohlwend, D, Gerhardt, S, Gnandt, E, Friedrich, T. | | Deposit date: | 2018-12-18 | | Release date: | 2019-06-26 | | Last modified: | 2024-05-15 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | A mechanism to prevent production of reactive oxygen species by Escherichia coli respiratory complex I.
Nat Commun, 10, 2019
|
|
6Q9J
 
 | | Crystal structure of reduced Aquifex aeolicus NADH-quinone oxidoreductase subunits NuoE G129S and NuoF bound to NADH | | Descriptor: | 1,4-DIHYDRONICOTINAMIDE ADENINE DINUCLEOTIDE, FE2/S2 (INORGANIC) CLUSTER, FLAVIN MONONUCLEOTIDE, ... | | Authors: | Wohlwend, D, Gerhardt, S, Gnandt, E, Friedrich, T. | | Deposit date: | 2018-12-18 | | Release date: | 2019-06-26 | | Last modified: | 2024-05-15 | | Method: | X-RAY DIFFRACTION (1.83 Å) | | Cite: | A mechanism to prevent production of reactive oxygen species by Escherichia coli respiratory complex I.
Nat Commun, 10, 2019
|
|
