8G5S
 
 | | Crystal structure of apo TnmJ | | Descriptor: | TnmJ | | Authors: | Liu, Y.-C, Li, G, Gui, C, Shen, B. | | Deposit date: | 2023-02-14 | | Release date: | 2023-10-18 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | Cofactorless oxygenases guide anthraquinone-fused enediyne biosynthesis.
Nat.Chem.Biol., 20, 2024
|
|
3COC
 
 | |
6DUP
 
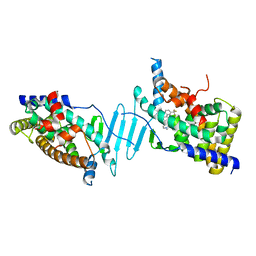 | | CRYSTAL STRUCTURE OF PXR IN COMPLEX WITH COMPOUND 7 | | Descriptor: | (2S)-2-({[3'-(trifluoromethyl)[1,1'-biphenyl]-4-yl]oxy}methyl)-2,3-dihydro-7H-[1,3]oxazolo[3,2-a]pyrimidin-7-one, Nuclear receptor subfamily 1 group I member 2 | | Authors: | Chen, X, Zhang, Y, Mclean, L.R. | | Deposit date: | 2018-06-21 | | Release date: | 2018-08-29 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Amelioration of PXR-mediated CYP3A4 induction by mGluR2 modulators.
Bioorg. Med. Chem. Lett., 28, 2018
|
|
6DA7
 
 | | Crystal structure of the TtnD decarboxylase from the tautomycetin biosynthesis pathway of Streptomyces griseochromogenes with apo form at 1.83 A resolution (I222) | | Descriptor: | 4-(2-HYDROXYETHYL)-1-PIPERAZINE ETHANESULFONIC ACID, GLYCEROL, SODIUM ION, ... | | Authors: | Han, L, Rudolf, J.D, Chang, C.-Y, Miller, M.D, Soman, J, Phillips Jr, G.N, Shen, B, Enzyme Discovery for Natural Product Biosynthesis (NatPro) | | Deposit date: | 2018-05-01 | | Release date: | 2018-10-03 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (1.83 Å) | | Cite: | Biochemical and Structural Characterization of TtnD, a Prenylated FMN-Dependent Decarboxylase from the Tautomycetin Biosynthetic Pathway.
ACS Chem. Biol., 13, 2018
|
|
6DA6
 
 | | Crystal structure of the TtnD decarboxylase from the tautomycetin biosynthesis pathway of Streptomyces griseochromogenes, apo form at 2.6 A resolution (P212121) | | Descriptor: | GLYCEROL, MAGNESIUM ION, UNKNOWN LIGAND, ... | | Authors: | Han, L, Rudolf, J.D, Chang, C.-Y, Miller, M.D, Soman, J, Phillips Jr, G.N, Shen, B, Enzyme Discovery for Natural Product Biosynthesis (NatPro) | | Deposit date: | 2018-05-01 | | Release date: | 2018-10-03 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (2.59 Å) | | Cite: | Biochemical and Structural Characterization of TtnD, a Prenylated FMN-Dependent Decarboxylase from the Tautomycetin Biosynthetic Pathway.
ACS Chem. Biol., 13, 2018
|
|
8ITM
 
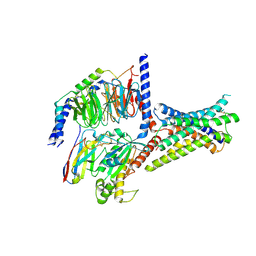 | | Cryo-EM structure of GIPR splice variant 2 (SV2) in complex with Gs protein | | Descriptor: | Gastric inhibitory polypeptide receptor, Guanine nucleotide-binding protein G(I)/G(S)/G(O) subunit gamma-2, Guanine nucleotide-binding protein G(I)/G(S)/G(T) subunit beta-1, ... | | Authors: | Zhao, F.H, Hang, K.N, Zhou, Q.T, Shao, L.J, Li, H, Li, W.Z, Lin, S, Dai, A.T, Cai, X.Q, Liu, Y.Y, Xu, Y.N, Feng, W.B, Yang, D.H, Wang, M.W. | | Deposit date: | 2023-03-22 | | Release date: | 2023-10-18 | | Method: | ELECTRON MICROSCOPY (3.13 Å) | | Cite: | Molecular basis of signal transduction mediated by the human GIPR splice variants.
Proc.Natl.Acad.Sci.USA, 120, 2023
|
|
8ITL
 
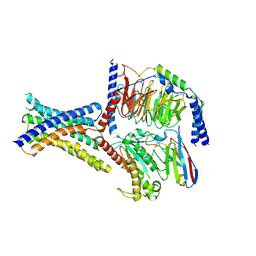 | | Cryo-EM structure of GIPR splice variant 1 (SV1) in complex with Gs protein | | Descriptor: | Gastric inhibitory polypeptide receptor, Guanine nucleotide-binding protein G(I)/G(S)/G(O) subunit gamma-2, Guanine nucleotide-binding protein G(I)/G(S)/G(T) subunit beta-1, ... | | Authors: | Zhao, F.H, Hang, K.N, Zhou, Q.T, Shao, L.J, Li, H, Li, W.Z, Lin, S, Dai, A.T, Cai, X.Q, Liu, Y.Y, Xu, Y.N, Feng, W.B, Yang, D.H, Wang, M.W. | | Deposit date: | 2023-03-22 | | Release date: | 2023-10-18 | | Method: | ELECTRON MICROSCOPY (3.23 Å) | | Cite: | Molecular basis of signal transduction mediated by the human GIPR splice variants.
Proc.Natl.Acad.Sci.USA, 120, 2023
|
|
5GK2
 
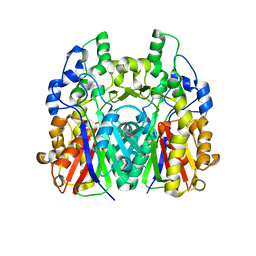 | |
7CU6
 
 | | lasso peptide C24 mutant - A11V2C | | Descriptor: | lasso peptide C24_A11V2C | | Authors: | Ma, M, Liu, X.H. | | Deposit date: | 2020-08-21 | | Release date: | 2021-08-25 | | Last modified: | 2023-06-14 | | Method: | SOLUTION NMR | | Cite: | Rational generation of lasso peptides based on biosynthetic gene mutations and site-selective chemical modifications.
Chem Sci, 12, 2021
|
|
5GK0
 
 | |
5GK1
 
 | | Crystal structure of the ketosynthase StlD complexed with substrate | | Descriptor: | 3-OXO-5-METHYLHEXANOIC ACID, Ketosynthase StlD | | Authors: | Mori, T, Saito, Y, Morita, H, Abe, I. | | Deposit date: | 2016-07-03 | | Release date: | 2017-07-05 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (1.83 Å) | | Cite: | Structural Insight into the Enzymatic Formation of Bacterial Stilbene.
Cell Chem Biol, 23, 2016
|
|
8IOD
 
 | | Cryo-EM structure of the PG-901-bound human melanocortin receptor 5 (MC5R)-Gs complex | | Descriptor: | CALCIUM ION, Guanine nucleotide-binding protein G(I)/G(S)/G(O) subunit gamma-2, Guanine nucleotide-binding protein G(I)/G(S)/G(T) subunit beta-1,HiBiT, ... | | Authors: | Feng, W.B, Zhou, Q.T, Chen, X.Y, Dai, A.T, Cai, X.Q, Liu, X, Zhao, F.H, Chen, Y, Ye, C.Y, Xu, Y.N, Cong, Z.T, Li, H, Lin, S, Yang, D.H, Wang, M.W. | | Deposit date: | 2023-03-10 | | Release date: | 2023-09-20 | | Last modified: | 2023-11-15 | | Method: | ELECTRON MICROSCOPY (2.59 Å) | | Cite: | Structural insights into ligand recognition and subtype selectivity of the human melanocortin-3 and melanocortin-5 receptors.
Cell Discov, 9, 2023
|
|
8INR
 
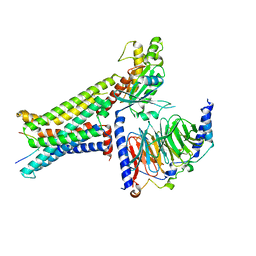 | | Cryo-EM structure of the alpha-MSH-bound human melanocortin receptor 5 (MC5R)-Gs complex | | Descriptor: | CALCIUM ION, Guanine nucleotide-binding protein G(I)/G(S)/G(O) subunit gamma-2, Guanine nucleotide-binding protein G(I)/G(S)/G(T) subunit beta-1,HiBiT, ... | | Authors: | Feng, W.B, Zhou, Q.T, Chen, X.Y, Dai, A.T, Cai, X.Q, Liu, X, Zhao, F.H, Chen, Y, Ye, C.Y, Xu, Y.N, Cong, Z.T, Li, H, Lin, S, Yang, D.H, Wang, M.W. | | Deposit date: | 2023-03-10 | | Release date: | 2023-09-20 | | Method: | ELECTRON MICROSCOPY (2.73 Å) | | Cite: | Structural insights into ligand recognition and subtype selectivity of the human melanocortin-3 and melanocortin-5 receptors.
Cell Discov, 9, 2023
|
|
8E18
 
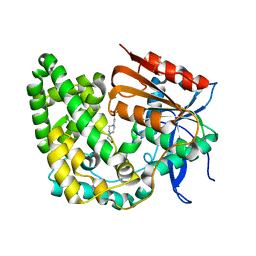 | | Crystal structure of apo TnmK1 | | Descriptor: | 4-(2-HYDROXYETHYL)-1-PIPERAZINE ETHANESULFONIC ACID, Secreted hydrolase | | Authors: | Liu, Y.-C, Gui, C, Shen, B. | | Deposit date: | 2022-08-10 | | Release date: | 2022-11-09 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (1.14 Å) | | Cite: | Intramolecular C-C Bond Formation Links Anthraquinone and Enediyne Scaffolds in Tiancimycin Biosynthesis.
J.Am.Chem.Soc., 144, 2022
|
|
8E19
 
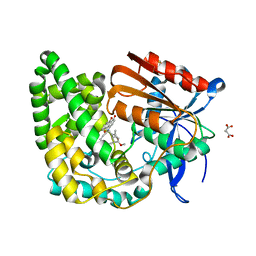 | | Crystal structure of TnmK1 complexed with TNM H | | Descriptor: | (1R,8S,13S)-8-[(4-hydroxy-9,10-dioxo-9,10-dihydroanthracen-1-yl)amino]-12-methoxy-10-methylbicyclo[7.3.1]trideca-9,11-diene-2,6-diyne-13-carbaldehyde, 4-(2-HYDROXYETHYL)-1-PIPERAZINE ETHANESULFONIC ACID, SUCCINIC ACID, ... | | Authors: | Liu, Y.-C, Gui, C, Shen, B. | | Deposit date: | 2022-08-10 | | Release date: | 2022-11-09 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (2.03 Å) | | Cite: | Intramolecular C-C Bond Formation Links Anthraquinone and Enediyne Scaffolds in Tiancimycin Biosynthesis.
J.Am.Chem.Soc., 144, 2022
|
|
1ZMQ
 
 | | Crystal structure of human alpha-defensin-6 | | Descriptor: | CHLORIDE ION, Defensin 6 | | Authors: | Lubkowski, J, Szyk, A, Lu, W. | | Deposit date: | 2005-05-10 | | Release date: | 2006-05-30 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Crystal structures of human {alpha}-defensins HNP4, HD5, and HD6.
Protein Sci., 15, 2006
|
|
6BBX
 
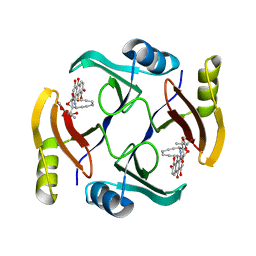 | | Crystal structure of TnmS3 in complex with TNM C | | Descriptor: | Glyoxalase/bleomycin resisance protein/dioxygenase, methyl (2R,3R)-2,3-dihydroxy-3-[(1aS,11S,11aR,14Z,18R)-3,7,8,18-tetrahydroxy-4,9-dioxo-4,9,10,11-tetrahydro-11aH-11,1a-hept[3]ene[1,5]diynonaphtho[2,3-h]oxireno[c]quinolin-11a-yl]butanoate | | Authors: | Chang, C.Y, Chang, C, Nocek, B, Rudolf, J.D, Joachimiak, A, Phillips Jr, G.N, Shen, B, Enzyme Discovery for Natural Product Biosynthesis (NatPro), Midwest Center for Structural Genomics (MCSG) | | Deposit date: | 2017-10-19 | | Release date: | 2018-07-04 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Resistance to Enediyne Antitumor Antibiotics by Sequestration.
Cell Chem Biol, 25, 2018
|
|
6ILQ
 
 | | Crystal structure of PPARgamma with compound BR101549 | | Descriptor: | Nuclear receptor coactivator 1, Peroxisome proliferator-activated receptor gamma, ethyl [2-butyl-6-oxo-1-{[2'-(5-oxo-4,5-dihydro-1,2,4-oxadiazol-3-yl)[1,1'-biphenyl]-4-yl]methyl}-4-(propan-2-yl)-1,6-dihydropyrimidin-5-yl]acetate | | Authors: | Hong, E, Jang, T.H, Chin, J, Kim, K.H, Jung, W, Kim, S.H. | | Deposit date: | 2018-10-19 | | Release date: | 2019-09-11 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (2.408 Å) | | Cite: | Identification of BR101549 as a lead candidate of non-TZD PPAR gamma agonist for the treatment of type 2 diabetes: Proof-of-concept evaluation and SAR.
Bioorg.Med.Chem.Lett., 29, 2019
|
|
5WUG
 
 | | Expression, characterization and crystal structure of a novel beta-glucosidase from Paenibacillus barengoltzii | | Descriptor: | Beta-glucosidase | | Authors: | Jiang, Z, Wu, S, Yang, D, Qin, Z, You, X, Huang, P. | | Deposit date: | 2016-12-17 | | Release date: | 2018-01-31 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (2.216 Å) | | Cite: | Expression, Biochemical Characterization and Structure Resolution of beta-glucosidase from Paenibacillus barengoltzii
J Food Sci Technol(China), 2019
|
|
1ZMP
 
 | | Crystal structure of human defensin-5 | | Descriptor: | CHLORIDE ION, Defensin 5, GLYCEROL, ... | | Authors: | Lubkowski, J, Szyk, A, Lu, W. | | Deposit date: | 2005-05-10 | | Release date: | 2006-05-30 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (1.65 Å) | | Cite: | Crystal structures of human {alpha}-defensins HNP4, HD5, and HD6.
Protein Sci., 15, 2006
|
|
1Y6X
 
 | |
6CLX
 
 | | Crystal structure of TnmH in complex with SAM | | Descriptor: | O-methyltransferase, S-ADENOSYLMETHIONINE | | Authors: | Chang, C.Y, Annaval, T, Adhikari, A, Yan, X, Shen, B. | | Deposit date: | 2018-03-02 | | Release date: | 2019-03-06 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (2.73 Å) | | Cite: | Characterization of TnmH as anO-Methyltransferase Revealing Insights into Tiancimycin Biosynthesis and Enabling a Biocatalytic Strategy To Prepare Antibody-Tiancimycin Conjugates.
J.Med.Chem., 63, 2020
|
|
1ZMM
 
 | |
6CLW
 
 | | Crystal structure of TnmH | | Descriptor: | O-methyltransferase | | Authors: | Chang, C.Y, Annaval, T, Adhikari, A, Yan, X, Shen, B. | | Deposit date: | 2018-03-02 | | Release date: | 2019-03-06 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (2.74 Å) | | Cite: | Characterization of TnmH as anO-Methyltransferase Revealing Insights into Tiancimycin Biosynthesis and Enabling a Biocatalytic Strategy To Prepare Antibody-Tiancimycin Conjugates.
J.Med.Chem., 63, 2020
|
|
7BUT
 
 | |
