2ORS
 
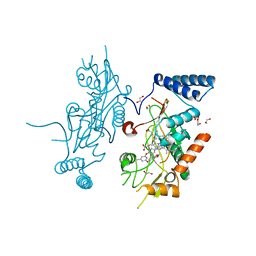 | | Murine Inducible Nitric Oxide Synthase Oxygenase Domain (DELTA 114) 4-(Benzo[1,3]dioxol-5-yloxy)-2-(4-imidazol-1-yl-phenoxy)-6-methyl-pyrimidine Complex | | Descriptor: | 1,2-ETHANEDIOL, 4-(1,3-BENZODIOXOL-5-YLOXY)-2-[4-(1H-IMIDAZOL-1-YL)PHENOXY]-6-METHYLPYRIMIDINE, Nitric oxide synthase, ... | | Authors: | Adler, M, Whitlow, M. | | Deposit date: | 2007-02-04 | | Release date: | 2007-04-17 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Design, Synthesis, and Activity of 2-Imidazol-1-ylpyrimidine Derived Inducible Nitric Oxide Synthase Dimerization Inhibitors
J.Med.Chem., 50, 2007
|
|
3B6Z
 
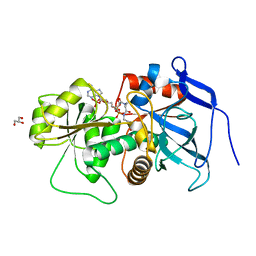 | | Lovastatin polyketide enoyl reductase (LovC) complexed with 2'-phosphoadenosyl isomer of crotonoyl-CoA | | Descriptor: | Enoyl reductase, GLYCEROL, S-{(9R,13R,15S)-17-[(2R,3R,4R,5R)-5-(6-amino-9H-purin-9-yl)-3-hydroxy-4-(phosphonooxy)tetrahydrofuran-2-yl]-9,13,15-trihydroxy-10,10-dimethyl-13,15-dioxido-4,8-dioxo-12,14,16-trioxa-3,7-diaza-13,15-diphosphaheptadec-1-yl}(2E)-but-2-enethioate | | Authors: | Ames, B.D, Smith, P.T, Ma, S.M, Wong, E.W, Xie, X, Vederas, J.C, Tang, Y, Tsai, S.-C. | | Deposit date: | 2007-10-29 | | Release date: | 2008-09-16 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (1.88 Å) | | Cite: | Crystal structure and biochemical studies of the trans-acting polyketide enoyl reductase LovC from lovastatin biosynthesis.
Proc.Natl.Acad.Sci.USA, 109, 2012
|
|
5LP8
 
 | |
6PTZ
 
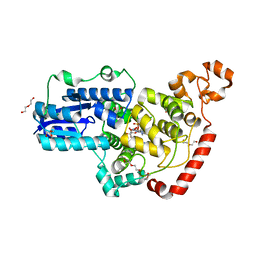 | | Crystal structure of pigeon Cryptochrome 4 mutant Y319D in complex with flavin adenine dinucleotide | | Descriptor: | Cryptochrome-1, DI(HYDROXYETHYL)ETHER, FLAVIN-ADENINE DINUCLEOTIDE, ... | | Authors: | Zoltowski, B.D, Chelliah, Y, Wickramaratne, A.C, Jarocha, L, Karki, N, Mouritsen, H, Hore, P.J, Hibbs, R.E, Green, C.B, Takahashi, J.S. | | Deposit date: | 2019-07-16 | | Release date: | 2019-09-04 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (1.793 Å) | | Cite: | Chemical and structural analysis of a photoactive vertebrate cryptochrome from pigeon.
Proc.Natl.Acad.Sci.USA, 116, 2019
|
|
6PU0
 
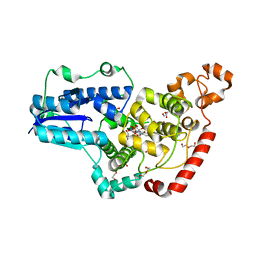 | | Pigeon Cryptochrome4 bound to flavin adenine dinucleotide | | Descriptor: | 1,2-ETHANEDIOL, Cryptochrome-1, DI(HYDROXYETHYL)ETHER, ... | | Authors: | Zoltowski, B.D, Chelliah, Y, Wickramaratne, A.C, Jarocha, L, Karki, N, Mouritsen, H, Hore, P.J, Hibbs, R.E, Green, C.B, Takahashi, J.S. | | Deposit date: | 2019-07-16 | | Release date: | 2019-09-04 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (1.8979 Å) | | Cite: | Chemical and structural analysis of a photoactive vertebrate cryptochrome from pigeon.
Proc.Natl.Acad.Sci.USA, 116, 2019
|
|
4FF5
 
 | |
8JVD
 
 | |
8JUC
 
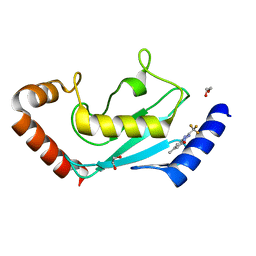 | | Identification of small-molecule binding sites of a ubiquitin-conjugating enzyme-UBE2T through fragment-based screening | | Descriptor: | 1,2-ETHANEDIOL, 7-methyl-2-(trifluoromethyl)-3~{H}-[1,2,4]triazolo[1,5-a]pyridin-5-one, Ubiquitin-conjugating enzyme E2 T | | Authors: | Anantharajan, J, Baburajendran, N. | | Deposit date: | 2023-06-26 | | Release date: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (1.54 Å) | | Cite: | Identification of small-molecule binding sites of a ubiquitin-conjugating enzyme-UBE2T through fragment-based screening.
Protein Sci., 33, 2024
|
|
8JRN
 
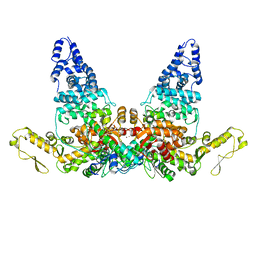 | | Structure of E6AP-E6 complex in Att1 state | | Descriptor: | Protein E6, Ubiquitin-protein ligase E3A, ZINC ION | | Authors: | Wang, Z, Yu, X. | | Deposit date: | 2023-06-17 | | Release date: | 2024-06-05 | | Method: | ELECTRON MICROSCOPY (2.6 Å) | | Cite: | Structural insights into the functional mechanism of the ubiquitin ligase E6AP.
Nat Commun, 15, 2024
|
|
8JRQ
 
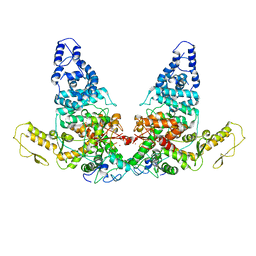 | | Structure of E6AP-E6 complex in Det1 state | | Descriptor: | Protein E6, Ubiquitin-protein ligase E3A, ZINC ION | | Authors: | Wang, Z, Yu, X. | | Deposit date: | 2023-06-17 | | Release date: | 2024-06-05 | | Method: | ELECTRON MICROSCOPY (4.15 Å) | | Cite: | Structural insights into the functional mechanism of the ubiquitin ligase E6AP.
Nat Commun, 15, 2024
|
|
8JRP
 
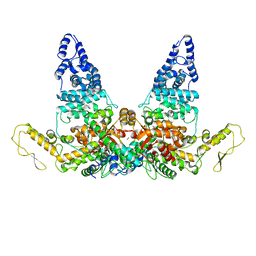 | | Structure of E6AP-E6 complex in Att3 state | | Descriptor: | Protein E6, Ubiquitin-protein ligase E3A, ZINC ION | | Authors: | Wang, Z, Yu, X. | | Deposit date: | 2023-06-17 | | Release date: | 2024-06-05 | | Method: | ELECTRON MICROSCOPY (3.58 Å) | | Cite: | Structural insights into the functional mechanism of the ubiquitin ligase E6AP.
Nat Commun, 15, 2024
|
|
8JRR
 
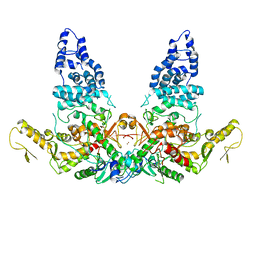 | | Structure of E6AP-E6 complex in Det2 state | | Descriptor: | Protein E6, Ubiquitin-protein ligase E3A, ZINC ION | | Authors: | Wang, Z, Yu, X. | | Deposit date: | 2023-06-17 | | Release date: | 2024-06-05 | | Method: | ELECTRON MICROSCOPY (4.35 Å) | | Cite: | Structural insights into the functional mechanism of the ubiquitin ligase E6AP.
Nat Commun, 15, 2024
|
|
8JRO
 
 | | Structure of E6AP-E6 complex in Att2 state | | Descriptor: | Protein E6, Ubiquitin-protein ligase E3A, ZINC ION | | Authors: | Wang, Z, Yu, X. | | Deposit date: | 2023-06-17 | | Release date: | 2024-06-05 | | Method: | ELECTRON MICROSCOPY (3.01 Å) | | Cite: | Structural insights into the functional mechanism of the ubiquitin ligase E6AP.
Nat Commun, 15, 2024
|
|
8K2S
 
 | |
8K2R
 
 | |
8K2T
 
 | |
7BTF
 
 | | SARS-CoV-2 RNA-dependent RNA polymerase in complex with cofactors in reduced condition | | Descriptor: | Non-structural protein 7, Non-structural protein 8, RNA-directed RNA polymerase, ... | | Authors: | Gao, Y, Yan, L, Huang, Y, Liu, F, Cao, L, Wang, T, Wang, Q, Lou, Z, Rao, Z. | | Deposit date: | 2020-04-01 | | Release date: | 2020-04-08 | | Last modified: | 2024-03-27 | | Method: | ELECTRON MICROSCOPY (2.95 Å) | | Cite: | Structure of the RNA-dependent RNA polymerase from COVID-19 virus.
Science, 368, 2020
|
|
7BBG
 
 | | CRYSTAL STRUCTURE OF HLA-A2-WT1-RMF AND FAB 11D06 | | Descriptor: | Beta-2-microglobulin, Heavy chain of Fab fragment 11D06, Light chain of Fab fragment 11D06, ... | | Authors: | Bujotzek, A, Georges, G, Hanisch, L.J, Klein, C, Benz, J. | | Deposit date: | 2020-12-17 | | Release date: | 2021-10-27 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (2.64 Å) | | Cite: | Targeting intracellular WT1 in AML with a novel RMF-peptide-MHC-specific T-cell bispecific antibody.
Blood, 138, 2021
|
|
6M71
 
 | | SARS-Cov-2 RNA-dependent RNA polymerase in complex with cofactors | | Descriptor: | Non-structural protein 7, Non-structural protein 8, RNA-directed RNA polymerase | | Authors: | Gao, Y, Yan, L, Huang, Y, Liu, F, Cao, L, Wang, T, Wang, Q, Lou, Z, Rao, Z. | | Deposit date: | 2020-03-16 | | Release date: | 2020-04-01 | | Last modified: | 2024-10-16 | | Method: | ELECTRON MICROSCOPY (2.9 Å) | | Cite: | Structure of the RNA-dependent RNA polymerase from COVID-19 virus.
Science, 368, 2020
|
|
6LXT
 
 | | Structure of post fusion core of 2019-nCoV S2 subunit | | Descriptor: | Spike protein S2, TETRAETHYLENE GLYCOL, ZINC ION | | Authors: | Zhu, Y, Sun, F. | | Deposit date: | 2020-02-11 | | Release date: | 2020-02-26 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2.9 Å) | | Cite: | Inhibition of SARS-CoV-2 (previously 2019-nCoV) infection by a highly potent pan-coronavirus fusion inhibitor targeting its spike protein that harbors a high capacity to mediate membrane fusion.
Cell Res., 30, 2020
|
|
7V2A
 
 | | SARS-CoV-2 Spike trimer in complex with XG014 Fab | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, Spike glycoprotein, The heavy chain of XG014, ... | | Authors: | Wang, K, Wang, X, Pan, L. | | Deposit date: | 2021-08-07 | | Release date: | 2021-10-20 | | Last modified: | 2024-10-16 | | Method: | ELECTRON MICROSCOPY (3.4 Å) | | Cite: | An ultrapotent pan-beta-coronavirus lineage B ( beta-CoV-B) neutralizing antibody locks the receptor-binding domain in closed conformation by targeting its conserved epitope.
Protein Cell, 13, 2022
|
|
7V26
 
 | | XG005-bound SARS-CoV-2 S | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, Spike glycoprotein, XG005 Heavy chain, ... | | Authors: | Zhan, W.Q, Zhang, X, Sun, L, Chen, Z.G. | | Deposit date: | 2021-08-07 | | Release date: | 2021-10-20 | | Last modified: | 2024-10-16 | | Method: | ELECTRON MICROSCOPY (3.85 Å) | | Cite: | An ultrapotent pan-beta-coronavirus lineage B ( beta-CoV-B) neutralizing antibody locks the receptor-binding domain in closed conformation by targeting its conserved epitope.
Protein Cell, 13, 2022
|
|
6NZN
 
 | | Dimer-of-dimer amyloid fibril structure of glucagon | | Descriptor: | Glucagon | | Authors: | Gelenter, M.D, Smith, K.J, Liao, S.Y, Mandala, V.S, Dregni, A.J, Lamm, M.S, Tian, Y, Wei, X, Pochan, D.J, Tucker, T.J, Su, Y, Hong, M. | | Deposit date: | 2019-02-14 | | Release date: | 2019-06-05 | | Last modified: | 2024-05-15 | | Method: | SOLID-STATE NMR | | Cite: | The peptide hormone glucagon forms amyloid fibrils with two coexisting beta-strand conformations.
Nat.Struct.Mol.Biol., 26, 2019
|
|
1OP2
 
 | | Crystal Structure of AaV-SP-II, a Glycosylated Snake Venom Serine Proteinase from Agkistrodon acutus | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, SULFATE ION, Venom serine proteinase | | Authors: | Zhu, Z, Teng, M, Niu, L. | | Deposit date: | 2003-03-04 | | Release date: | 2004-05-25 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Crystal Structures and Amidolytic Activities of Two Glycosylated Snake Venom Serine Proteinases
J.BIOL.CHEM., 280, 2005
|
|
5ZUV
 
 | | Crystal Structure of the Human Coronavirus 229E HR1 motif in complex with pan-CoVs inhibitor EK1 | | Descriptor: | CHLORIDE ION, Spike glycoprotein,Spike glycoprotein,inhibitor EK1 | | Authors: | Yan, L, Yang, B, Wilson, I.A. | | Deposit date: | 2018-05-08 | | Release date: | 2019-04-10 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (2.21 Å) | | Cite: | A pan-coronavirus fusion inhibitor targeting the HR1 domain of human coronavirus spike.
Sci Adv, 5, 2019
|
|
