7LRD
 
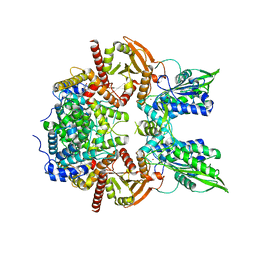 | | Cryo-EM of the SLFN12-PDE3A complex: Consensus subset model | | Descriptor: | (4~{R})-3-[4-(diethylamino)-3-[oxidanyl(oxidanylidene)-$l^{4}-azanyl]phenyl]-4-methyl-4,5-dihydro-1~{H}-pyridazin-6-one, MAGNESIUM ION, MANGANESE (II) ION, ... | | Authors: | Fuller, J.R, Garvie, C.W, Lemke, C.T. | | Deposit date: | 2021-02-16 | | Release date: | 2021-06-09 | | Last modified: | 2024-03-06 | | Method: | ELECTRON MICROSCOPY (3.22 Å) | | Cite: | Structure of PDE3A-SLFN12 complex reveals requirements for activation of SLFN12 RNase.
Nat Commun, 12, 2021
|
|
7LRE
 
 | |
7LRC
 
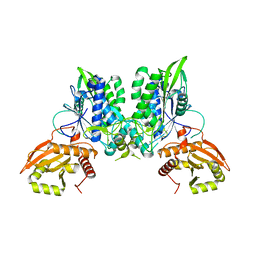
 | | Cryo-EM of the SLFN12-PDE3A complex: PDE3A body refinement | | Descriptor: | (4~{R})-3-[4-(diethylamino)-3-[oxidanyl(oxidanylidene)-$l^{4}-azanyl]phenyl]-4-methyl-4,5-dihydro-1~{H}-pyridazin-6-one, MAGNESIUM ION, MANGANESE (II) ION, ... | | Authors: | Fuller, J.R, Garvie, C.W, Lemke, C.T. | | Deposit date: | 2021-02-16 | | Release date: | 2021-06-09 | | Last modified: | 2024-03-06 | | Method: | ELECTRON MICROSCOPY (2.97 Å) | | Cite: | Structure of PDE3A-SLFN12 complex reveals requirements for activation of SLFN12 RNase.
Nat Commun, 12, 2021
|
|
7L28
 
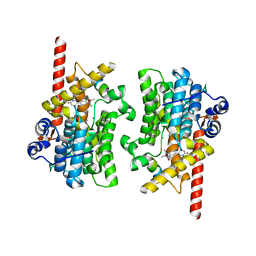
 | | Crystal structure of the catalytic domain of human PDE3A bound to Trequinsin | | Descriptor: | (2E)-9,10-dimethoxy-3-methyl-2-[(2,4,6-trimethylphenyl)imino]-2,3,6,7-tetrahydro-4H-pyrimido[6,1-a]isoquinolin-4-one, ACETATE ION, MAGNESIUM ION, ... | | Authors: | Horner, S.W, Garvie, C. | | Deposit date: | 2020-12-16 | | Release date: | 2021-06-16 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Structure of PDE3A-SLFN12 complex reveals requirements for activation of SLFN12 RNase.
Nat Commun, 12, 2021
|
|
7L29
 
 | |
7L27
 
 | |
5HYZ
 
 | | Crystal Structure of SCL7 in Oryza sativa | | Descriptor: | GRAS family transcription factor containing protein, expressed | | Authors: | Wu, Y, Li, S, Zhao, Y, Sun, L. | | Deposit date: | 2016-02-02 | | Release date: | 2016-04-27 | | Last modified: | 2024-11-20 | | Method: | X-RAY DIFFRACTION (1.822 Å) | | Cite: | Crystal Structure of the GRAS Domain of SCARECROW-LIKE7 in Oryza sativa.
Plant Cell, 28, 2016
|
|
7KWE
 
 | | Crystal structure of the catalytic domain of human PDE3A bound to DNMDP | | Descriptor: | (4~{R})-3-[4-(diethylamino)-3-[oxidanyl(oxidanylidene)-$l^{4}-azanyl]phenyl]-4-methyl-4,5-dihydro-1~{H}-pyridazin-6-one, ACETATE ION, MAGNESIUM ION, ... | | Authors: | Horner, S.W, Garvie, C. | | Deposit date: | 2020-11-30 | | Release date: | 2021-06-16 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Structure of PDE3A-SLFN12 complex reveals requirements for activation of SLFN12 RNase.
Nat Commun, 12, 2021
|
|
9MOE
 
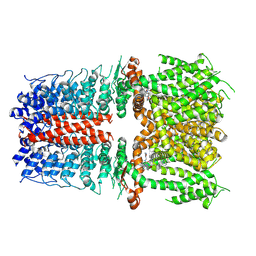 | | Preclinical and clinical evaluation of a novel TRPA1 antagonist LY3526318 | | Descriptor: | (2S)-2-{3-methyl-1-[(3-methyl-1,2,4-oxadiazol-5-yl)methyl]-2,6-dioxo-1,2,3,6-tetrahydro-7H-purin-7-yl}-N-{6-[2-(trifluoromethyl)pyrimidin-5-yl]pyridin-2-yl}propanamide, Transient receptor potential cation channel subfamily A member 1 | | Authors: | Nie, S. | | Deposit date: | 2024-12-26 | | Release date: | 2025-05-21 | | Method: | ELECTRON MICROSCOPY (2.7 Å) | | Cite: | Preclinical and clinical evaluation of a novel TRPA1 antagonist LY3526318.
Pain, 2025
|
|
7F0R
 
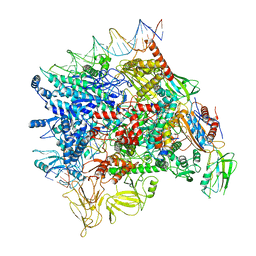 | | Cryo-EM structure of Pseudomonas aeruginosa SutA transcription activation complex | | Descriptor: | DNA (70-MER), DNA-directed RNA polymerase subunit alpha, DNA-directed RNA polymerase subunit beta, ... | | Authors: | He, D.W, You, L.L, Zhang, Y. | | Deposit date: | 2021-06-06 | | Release date: | 2022-07-27 | | Last modified: | 2024-06-12 | | Method: | ELECTRON MICROSCOPY (5.8 Å) | | Cite: | Pseudomonas aeruginosa SutA wedges RNAP lobe domain open to facilitate promoter DNA unwinding.
Nat Commun, 13, 2022
|
|
7Q9N
 
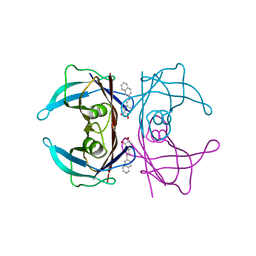 | | Transthyretin complexed with (E)-4-(2-(naphthalen-2-yl)vinyl)benzene-1,2-diol | | Descriptor: | 4-[(~{E})-2-naphthalen-2-ylethenyl]benzene-1,2-diol, Transthyretin | | Authors: | Derbyshire, D.J, Hammarstrom, P, von Castelmur, E, Begum, A. | | Deposit date: | 2021-11-12 | | Release date: | 2022-11-23 | | Last modified: | 2024-05-01 | | Method: | X-RAY DIFFRACTION (1.45 Å) | | Cite: | Transthyretin Binding Mode Dichotomy of Fluorescent trans -Stilbene Ligands.
Acs Chem Neurosci, 14, 2023
|
|
7Q9O
 
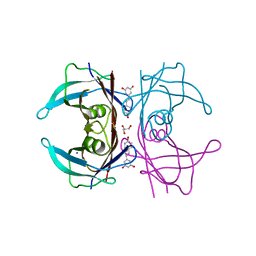 | | Complex of Transthyretin with resveratrol exhibits multiple binding modes | | Descriptor: | GLYCEROL, RESVERATROL, SODIUM ION, ... | | Authors: | Derbyshire, D.J, Hammarstrom, P, von Castelmur, E, Begum, A. | | Deposit date: | 2021-11-12 | | Release date: | 2022-11-23 | | Last modified: | 2024-05-01 | | Method: | X-RAY DIFFRACTION (1.35 Å) | | Cite: | Transthyretin Binding Mode Dichotomy of Fluorescent trans -Stilbene Ligands.
Acs Chem Neurosci, 14, 2023
|
|
7Q9L
 
 | | Transthyretin complexed with (E)-4-(2-(naphthalen-1-yl)vinyl)benzene-1,2-diol | | Descriptor: | 4-[(~{E})-2-naphthalen-1-ylethenyl]benzene-1,2-diol, GLYCEROL, SODIUM ION, ... | | Authors: | Derbyshire, D.J, Hammarstrom, P, von Castelmur, E, Begum, A. | | Deposit date: | 2021-11-12 | | Release date: | 2022-11-23 | | Last modified: | 2024-05-01 | | Method: | X-RAY DIFFRACTION (1.45 Å) | | Cite: | Transthyretin Binding Mode Dichotomy of Fluorescent trans -Stilbene Ligands.
Acs Chem Neurosci, 14, 2023
|
|
7RAI
 
 | |
7TJE
 
 | | Bacteriophage Q beta capsid protein A38K | | Descriptor: | Minor capsid protein A1 | | Authors: | Jin, X. | | Deposit date: | 2022-01-16 | | Release date: | 2023-01-25 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (2.799 Å) | | Cite: | Alternative Assembly of Q beta Virus-like Particles
To Be Published
|
|
7TJG
 
 | |
7TJM
 
 | |
7TJD
 
 | |
6IEG
 
 | | Crystal structure of human MTR4 | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, Exosome RNA helicase MTR4, MAGNESIUM ION | | Authors: | Chen, J.Y, Yun, C.H. | | Deposit date: | 2018-09-14 | | Release date: | 2019-04-03 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (3.55 Å) | | Cite: | NRDE2 negatively regulates exosome functions by inhibiting MTR4 recruitment and exosome interaction.
Genes Dev., 33, 2019
|
|
6IEH
 
 | | Crystal structures of the hMTR4-NRDE2 complex | | Descriptor: | ADENOSINE-5'-TRIPHOSPHATE, CHLORIDE ION, Exosome RNA helicase MTR4, ... | | Authors: | Chen, J.Y, Yun, C.H. | | Deposit date: | 2018-09-14 | | Release date: | 2019-04-03 | | Last modified: | 2024-11-20 | | Method: | X-RAY DIFFRACTION (2.892 Å) | | Cite: | NRDE2 negatively regulates exosome functions by inhibiting MTR4 recruitment and exosome interaction.
Genes Dev., 33, 2019
|
|
4ANV
 
 | | Complexes of PI3Kgamma with isoform selective inhibitors. | | Descriptor: | 2-{4-[(4'-METHOXYBIPHENYL-3-YL)SULFONYL]PIPERAZIN-1-YL}-3-(4-METHOXYPHENYL)PYRAZINE, PHOSPHATIDYLINOSITOL-4,5-BISPHOSPHATE 3-KINASE CATALYTIC SUBUNIT GAMMA ISOFORM, SULFATE ION | | Authors: | Foster, P.G, Lougheed, J.C. | | Deposit date: | 2012-03-22 | | Release date: | 2012-05-09 | | Last modified: | 2024-05-01 | | Method: | X-RAY DIFFRACTION (2.13 Å) | | Cite: | The Discovery of a Novel Series of Potent and Orally Bioavailable Phosphoinositide 3-Kinase Gamma Inhibitors
J.Med.Chem., 55, 2012
|
|
4ANX
 
 | | Complexes of PI3Kgamma with isoform selective inhibitors. | | Descriptor: | 5-{3-[(4-{3-[4-(1-methylethyl)phenyl]pyrazin-2-yl}piperazin-1-yl)sulfonyl]phenyl}pyrimidin-2-amine, PHOSPHATIDYLINOSITOL-4,5-BISPHOSPHATE 3-KINASE CATALYTIC SUBUNIT GAMMA ISOFORM | | Authors: | Foster, P.G, Lougheed, J.C. | | Deposit date: | 2012-03-22 | | Release date: | 2012-05-09 | | Last modified: | 2024-05-01 | | Method: | X-RAY DIFFRACTION (2.73 Å) | | Cite: | The Discovery of a Novel Series of Potent and Orally Bioavailable Phosphoinositide 3-Kinase Gamma Inhibitors
J.Med.Chem., 55, 2012
|
|
4AW5
 
 | | Complex of the EphB4 kinase domain with an oxindole inhibitor | | Descriptor: | (3Z)-5-[(1-ethylpiperidin-4-yl)amino]-3-[(5-methoxy-1H-benzimidazol-2-yl)(phenyl)methylidene]-1,3-dihydro-2H-indol-2-one, EPHRIN TYPE-B RECEPTOR 4 | | Authors: | Till, J.H, Stout, T.J. | | Deposit date: | 2012-05-31 | | Release date: | 2012-08-01 | | Last modified: | 2024-05-01 | | Method: | X-RAY DIFFRACTION (2.33 Å) | | Cite: | The Design, Synthesis, and Biological Evaluation of Potent Receptor Tyrosine Kinase Inhibitors.
Bioorg.Med.Chem.Lett., 22, 2012
|
|
6VU2
 
 | | M1214_N1 Fab structure | | Descriptor: | M1214 N1 Fab heavy chain, M1214 N1 Fab light chain | | Authors: | Pan, R, Kong, X. | | Deposit date: | 2020-02-14 | | Release date: | 2020-05-06 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (2.19 Å) | | Cite: | VSV-Displayed HIV-1 Envelope Identifies Broadly Neutralizing Antibodies Class-Switched to IgG and IgA.
Cell Host Microbe, 27, 2020
|
|
4ANU
 
 | | Complexes of PI3Kgamma with isoform selective inhibitors. | | Descriptor: | 3-AMINO-N-METHYL-6-[3-(1H-TETRAZOL-5-YL)PHENYL]PYRAZINE-2-CARBOXAMIDE, PHOSPHATIDYLINOSITOL-4,5-BISPHOSPHATE 3-KINASE CATALYTIC SUBUNIT GAMMA ISOFORM, SULFATE ION | | Authors: | Foster, P.G, Lougheed, J.C. | | Deposit date: | 2012-03-22 | | Release date: | 2012-05-09 | | Last modified: | 2024-05-01 | | Method: | X-RAY DIFFRACTION (2.81 Å) | | Cite: | The Discovery of a Novel Series of Potent and Orally Bioavailable Phosphoinositide 3-Kinase Gamma Inhibitors
J.Med.Chem., 55, 2012
|
|
