6NJS
 
 | | Stat3 Core in complex with compound SD36 | | Descriptor: | Signal transducer and activator of transcription 3, [(2-{[(5S,8S,10aR)-3-acetyl-8-({(2S)-5-amino-1-[(diphenylmethyl)amino]-1,5-dioxopentan-2-yl}carbamoyl)-6-oxodecahydropyrrolo[1,2-a][1,5]diazocin-5-yl]carbamoyl}-1H-indol-5-yl)(difluoro)methyl]phosphonic acid (non-preferred name) | | Authors: | Meagher, J.L, Stuckey, J.A. | | Deposit date: | 2019-01-04 | | Release date: | 2019-12-04 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | A Potent and Selective Small-Molecule Degrader of STAT3 Achieves Complete Tumor Regression In Vivo.
Cancer Cell, 36, 2019
|
|
6NUQ
 
 | | Stat3 Core in complex with compound SI109 | | Descriptor: | Signal transducer and activator of transcription 3, [(2-{[(5S,8S,10aR)-3-acetyl-8-({(2S)-5-amino-1-[(diphenylmethyl)amino]-1,5-dioxopentan-2-yl}carbamoyl)-6-oxodecahydropyrrolo[1,2-a][1,5]diazocin-5-yl]carbamoyl}-1H-indol-5-yl)(difluoro)methyl]phosphonic acid (non-preferred name) | | Authors: | Meagher, J.L, Stuckey, J.A. | | Deposit date: | 2019-02-01 | | Release date: | 2019-12-04 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (3.15 Å) | | Cite: | A Potent and Selective Small-Molecule Degrader of STAT3 Achieves Complete Tumor Regression In Vivo.
Cancer Cell, 36, 2019
|
|
5BTU
 
 | |
2OJU
 
 | | X-ray structure of complex of human cyclophilin J with cyclosporin A | | Descriptor: | CYCLOSPORIN A, PEPTIDYL-PROLYL CIS-TRANS ISOMERASE-LIKE 3 | | Authors: | Xia, Z, Huang, L. | | Deposit date: | 2007-01-14 | | Release date: | 2008-01-22 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Targeting Cyclophilin J, a Novel Peptidyl-Prolyl Isomerase, Can Induce Cellular G1/S Arrest and Repress the Growth of Hepatocellular Carcinoma
To be Published
|
|
2OK3
 
 | | X-ray structure of human cyclophilin J at 2.0 angstrom | | Descriptor: | NICKEL (II) ION, Peptidyl-prolyl cis-trans isomerase-like 3 | | Authors: | Xia, Z. | | Deposit date: | 2007-01-15 | | Release date: | 2008-01-22 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Targeting Cyclophilin J, a novel peptidyl-prolyl isomerase, can induce cellular G1/S arrest and repress the growth of Hepatocellular carcinoma
To be Published
|
|
5C90
 
 | | Staphylococcus aureus ClpP mutant - Y63A | | Descriptor: | (4S)-2-METHYL-2,4-PENTANEDIOL, ATP-dependent Clp protease proteolytic subunit | | Authors: | Ye, F, Liu, H, Zhang, J, Gan, J, Yang, C.-G. | | Deposit date: | 2015-06-26 | | Release date: | 2016-05-25 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (1.75 Å) | | Cite: | Characterization of Gain-of-Function Mutant Provides New Insights into ClpP Structure
Acs Chem.Biol., 11, 2016
|
|
3UJL
 
 | | Crystal structure of abscisic acid bound PYL2 in complex with type 2C protein phosphatase ABI2 | | Descriptor: | (2Z,4E)-5-[(1S)-1-hydroxy-2,6,6-trimethyl-4-oxocyclohex-2-en-1-yl]-3-methylpenta-2,4-dienoic acid, Abscisic acid receptor PYL2, MAGNESIUM ION, ... | | Authors: | Zhou, X.E, Soon, F.-F, Ng, L.-M, Kovach, A, Tan, M.H.E, Suino-Powell, K.M, He, Y, Xu, Y, Brunzelle, J.S, Li, J, Melcher, K, Xu, H.E. | | Deposit date: | 2011-11-07 | | Release date: | 2012-02-15 | | Last modified: | 2024-10-09 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Molecular mimicry regulates ABA signaling by SnRK2 kinases and PP2C phosphatases.
Science, 335, 2012
|
|
3UJG
 
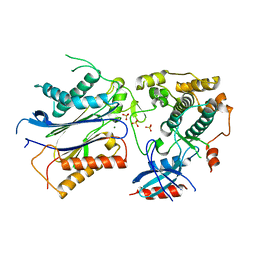 | | Crystal structure of SnRK2.6 in complex with HAB1 | | Descriptor: | MAGNESIUM ION, Protein phosphatase 2C 16, SULFATE ION, ... | | Authors: | Zhou, X.E, Soon, F.-F, Ng, L.-M, Kovach, A, Tan, M.H.E, Suino-Powell, K.M, He, Y, Xu, Y, Brunzelle, J.S, Li, J, Melcher, K, Xu, H.E. | | Deposit date: | 2011-11-07 | | Release date: | 2012-02-15 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Molecular mimicry regulates ABA signaling by SnRK2 kinases and PP2C phosphatases.
Science, 335, 2012
|
|
3UJK
 
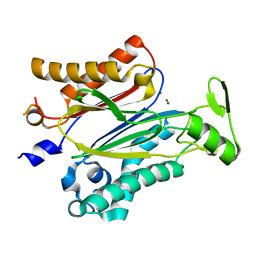 | | Crystal structure of protein phosphatase ABI2 | | Descriptor: | MAGNESIUM ION, Protein phosphatase 2C 77 | | Authors: | Zhou, X.E, Soon, F.-F, Ng, L.-M, Kovach, A, Tan, M.H.E, Suino-Powell, K.M, He, Y, Xu, Y, Brunzelle, J.S, Li, J, Melcher, K, Xu, H.E. | | Deposit date: | 2011-11-07 | | Release date: | 2012-02-15 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Molecular mimicry regulates ABA signaling by SnRK2 kinases and PP2C phosphatases.
Science, 335, 2012
|
|
1J4H
 
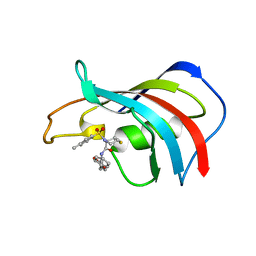 | | crystal structure analysis of the FKBP12 complexed with 000107 small molecule | | Descriptor: | 3-PHENYL-2-{[4-(TOLUENE-4-SULFONYL)-THIOMORPHOLINE-3-CARBONYL]-AMINO}-PROPIONIC ACID ETHYL ESTER, FKBP12 | | Authors: | Li, P, Ding, Y, Wang, L, Wu, B, Shu, C, Li, S, Shen, B, Rao, Z. | | Deposit date: | 2001-09-30 | | Release date: | 2003-06-03 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Design and structure-based study of new potential FKBP12 inhibitors.
Biophys.J., 85, 2003
|
|
8I2H
 
 | | Follicle stimulating hormone receptor | | Descriptor: | Follicle-stimulating hormone receptor | | Authors: | Duan, J, Xu, P, Yang, J, Ji, Y, Zhang, H, Mao, C, Luan, X, Jiang, Y, Zhang, Y, Zhang, S, Xu, H.E. | | Deposit date: | 2023-01-14 | | Release date: | 2023-03-22 | | Last modified: | 2024-07-03 | | Method: | ELECTRON MICROSCOPY (6 Å) | | Cite: | Mechanism of hormone and allosteric agonist mediated activation of follicle stimulating hormone receptor.
Nat Commun, 14, 2023
|
|
8I2G
 
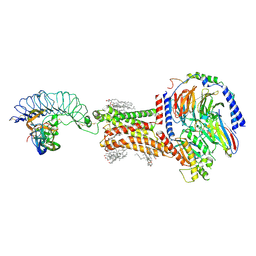 | | FSHR-Follicle stimulating hormone-compound 716340-Gs complex | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, CHOLESTEROL, ... | | Authors: | Duan, J, Xu, P, Yang, J, Ji, Y, Zhang, H, Mao, C, Luan, X, Jiang, Y, Zhang, Y, Zhang, S, Xu, H.E. | | Deposit date: | 2023-01-14 | | Release date: | 2023-03-29 | | Last modified: | 2024-11-20 | | Method: | ELECTRON MICROSCOPY (2.8 Å) | | Cite: | Mechanism of hormone and allosteric agonist mediated activation of follicle stimulating hormone receptor.
Nat Commun, 14, 2023
|
|
2NAE
 
 | | Membrane-bound mouse CD28 cytoplasmic tail | | Descriptor: | T-cell-specific surface glycoprotein CD28 | | Authors: | Li, H, Xu, C, Pan, W. | | Deposit date: | 2015-12-23 | | Release date: | 2016-12-21 | | Last modified: | 2024-05-15 | | Method: | SOLUTION NMR | | Cite: | Dynamic regulation of CD28 conformation and signaling by charged lipids and ions.
Nat.Struct.Mol.Biol., 24, 2017
|
|
1J4I
 
 | | crystal structure analysis of the FKBP12 complexed with 000308 small molecule | | Descriptor: | 4-METHYL-2-{[4-(TOLUENE-4-SULFONYL)-THIOMORPHOLINE-3-CARBONYL]-AMINO}-PENTANOIC ACID, FKBP12 | | Authors: | Li, P, Ding, Y, Wang, L, Wu, B, Shu, C, Li, S, Shen, B, Rao, Z. | | Deposit date: | 2001-09-30 | | Release date: | 2003-06-03 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Design and structure-based study of new potential FKBP12 inhibitors.
Biophys.J., 85, 2003
|
|
3UOB
 
 | |
2EW5
 
 | | Structure of Helicobacter Pylori peptide deformylase in complex with inhibitor | | Descriptor: | 4-{(1E)-3-OXO-3-[(2-PHENYLETHYL)AMINO]PROP-1-EN-1-YL}-1,2-PHENYLENE DIACETATE, COBALT (II) ION, peptide deformylase | | Authors: | Cai, J. | | Deposit date: | 2005-11-02 | | Release date: | 2006-10-24 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Peptide deformylase is a potential target for anti-Helicobacter pylori drugs: reverse docking, enzymatic assay, and X-ray crystallography validation
Protein Sci., 15, 2006
|
|
5BWN
 
 | | Crystal Structure of SIRT3 with a H3K9 Peptide Containing a Myristoyl Lysine | | Descriptor: | NAD-dependent protein deacetylase sirtuin-3, mitochondrial, ZINC ION, ... | | Authors: | Gai, W, Liu, D. | | Deposit date: | 2015-06-08 | | Release date: | 2016-07-13 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (1.942 Å) | | Cite: | Crystal structures of SIRT3 reveal that the alpha 2-alpha 3 loop and alpha 3-helix affect the interaction with long-chain acyl lysine.
Febs Lett., 590, 2016
|
|
3UO7
 
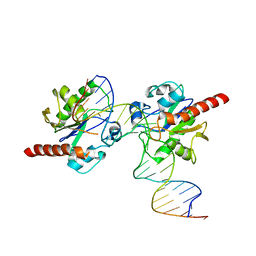 | | Crystal structure of Human Thymine DNA Glycosylase Bound to Substrate 5-carboxylcytosine | | Descriptor: | 5'-D(*CP*AP*GP*CP*TP*CP*TP*GP*TP*AP*CP*AP*TP*GP*AP*GP*CP*AP*GP*TP*GP*GP*A)-3', 5'-D(*CP*CP*AP*CP*TP*GP*CP*TP*CP*AP*(1CC)P*GP*TP*AP*CP*AP*GP*AP*GP*CP*TP*GP*T)-3', G/T mismatch-specific thymine DNA glycosylase | | Authors: | Zhang, L, He, C. | | Deposit date: | 2011-11-16 | | Release date: | 2012-02-15 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (3.002 Å) | | Cite: | Thymine DNA glycosylase specifically recognizes 5-carboxylcytosine-modified DNA.
Nat.Chem.Biol., 8, 2012
|
|
2EW7
 
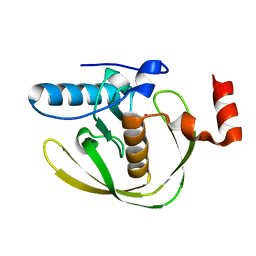 | | Crystal Structure of Helicobacter Pylori peptide deformylase | | Descriptor: | COBALT (II) ION, peptide deformylase | | Authors: | Cai, J. | | Deposit date: | 2005-11-02 | | Release date: | 2006-10-24 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Peptide deformylase is a potential target for anti-Helicobacter pylori drugs: reverse docking, enzymatic assay, and X-ray crystallography validation
Protein Sci., 15, 2006
|
|
2EW6
 
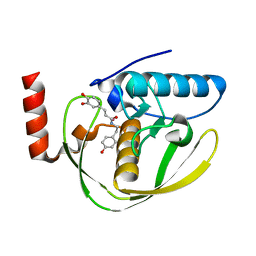 | | Structure of Helicobacter Pylori peptide deformylase in complex with inhibitor | | Descriptor: | (2E)-3-(3,4-DIHYDROXYPHENYL)-N-[2-(4-HYDROXYPHENYL)ETHYL]ACRYLAMIDE, COBALT (II) ION, peptide deformylase | | Authors: | Cai, J. | | Deposit date: | 2005-11-02 | | Release date: | 2006-10-24 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Peptide deformylase is a potential target for anti-Helicobacter pylori drugs: reverse docking, enzymatic assay, and X-ray crystallography validation
Protein Sci., 15, 2006
|
|
6CKH
 
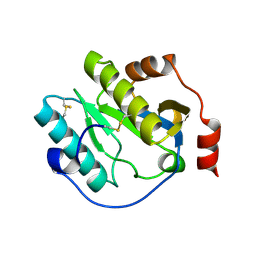 | | Manduca sexta Peptidoglycan Recognition Protein-1 | | Descriptor: | Peptidoglycan-recognition protein | | Authors: | Hu, Y. | | Deposit date: | 2018-02-28 | | Release date: | 2019-06-05 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | The three-dimensional structure and recognition mechanism of Manduca sexta peptidoglycan recognition protein-1.
Insect Biochem.Mol.Biol., 108, 2019
|
|
4IA0
 
 | | Crystal structure of the PDE5A1 catalytic domain in complex with novel inhibitors | | Descriptor: | 5-bromo-2-{2-ethoxy-5-[(4-methylpiperazin-1-yl)sulfonyl]phenyl}-6-octylpyrimidin-4(3H)-one, MAGNESIUM ION, ZINC ION, ... | | Authors: | Ren, J, Chen, T, Xu, Y. | | Deposit date: | 2012-12-05 | | Release date: | 2014-01-01 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (2.17 Å) | | Cite: | Exploration of the 5-bromopyrimidin-4(3H)-ones as potent inhibitors of PDE5.
Bioorg.Med.Chem.Lett., 23, 2013
|
|
4I9Z
 
 | | Crystal structure of the PDE5A1 catalytic domain in complex with novel inhibitors | | Descriptor: | 5-bromo-2-{5-[(4-methylpiperazin-1-yl)acetyl]-2-propoxyphenyl}-6-(propan-2-yl)pyrimidin-4(3H)-one, DI(HYDROXYETHYL)ETHER, MAGNESIUM ION, ... | | Authors: | Ren, J, Chen, T, Xu, Y. | | Deposit date: | 2012-12-05 | | Release date: | 2014-01-01 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (2.08 Å) | | Cite: | Exploration of the 5-bromopyrimidin-4(3H)-ones as potent inhibitors of PDE5.
Bioorg.Med.Chem.Lett., 23, 2013
|
|
6BTH
 
 | | Crystal structure of human cellular retinol binding protein 2 (CRBP2) in complex with 2-arachidonoylglycerol (2-AG) | | Descriptor: | 1,3-dihydroxypropan-2-yl (5Z,8Z,11Z,14Z)-icosa-5,8,11,14-tetraenoate, DI(HYDROXYETHYL)ETHER, Retinol-binding protein 2 | | Authors: | Silvaroli, J.A, Blaner, W.S, Lodowski, D.T, Golczak, M. | | Deposit date: | 2017-12-06 | | Release date: | 2018-12-12 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (1.35 Å) | | Cite: | Retinol-binding protein 2 (RBP2) binds monoacylglycerols and modulates gut endocrine signaling and body weight.
Sci Adv, 6, 2020
|
|
3B5H
 
 | |
