1ARW
 
 | | CRYSTAL STRUCTURES OF CYANIDE-AND TRIIODIDE-BOUND FORMS OF ARTHROMYCES RAMOSUS PEROXIDASE AT DIFFERENT PH VALUES. PERTURBATIONS OF ACTIVE SITE RESIDUES AND THEIR IMPLICATION IN ENZYME CATALYSIS | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, CALCIUM ION, CYANIDE ION, ... | | Authors: | Fukuyama, K, Kunishima, N, Amada, F. | | Deposit date: | 1995-04-25 | | Release date: | 1996-01-29 | | Last modified: | 2020-07-29 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Crystal structures of cyanide- and triiodide-bound forms of Arthromyces ramosus peroxidase at different pH values. Perturbations of active site residues and their implication in enzyme catalysis.
J.Biol.Chem., 270, 1995
|
|
1ARV
 
 | | CRYSTAL STRUCTURES OF CYANIDE-AND TRIIODIDE-BOUND FORMS OF ARTHROMYCES RAMOSUS PEROXIDASE AT DIFFERENT PH VALUES. PERTURBATIONS OF ACTIVE SITE RESIDUES AND THEIR IMPLICATION IN ENZYME CATALYSIS | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, CALCIUM ION, CYANIDE ION, ... | | Authors: | Fukuyama, K, Kunishima, N, Amada, F. | | Deposit date: | 1995-04-25 | | Release date: | 1996-01-29 | | Last modified: | 2020-07-29 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Crystal structures of cyanide- and triiodide-bound forms of Arthromyces ramosus peroxidase at different pH values. Perturbations of active site residues and their implication in enzyme catalysis.
J.Biol.Chem., 270, 1995
|
|
1ARU
 
 | | CRYSTAL STRUCTURES OF CYANIDE-AND TRIIODIDE-BOUND FORMS OF ARTHROMYCES RAMOSUS PEROXIDASE AT DIFFERENT PH VALUES. PERTURBATIONS OF ACTIVE SITE RESIDUES AND THEIR IMPLICATION IN ENZYME CATALYSIS | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, CALCIUM ION, CYANIDE ION, ... | | Authors: | Fukuyama, K, Kunishima, N, Amada, F. | | Deposit date: | 1995-04-25 | | Release date: | 1996-01-29 | | Last modified: | 2020-07-29 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Crystal structures of cyanide- and triiodide-bound forms of Arthromyces ramosus peroxidase at different pH values. Perturbations of active site residues and their implication in enzyme catalysis.
J.Biol.Chem., 270, 1995
|
|
1LAA
 
 | |
1ARX
 
 | | CRYSTAL STRUCTURES OF CYANIDE-AND TRIIODIDE-BOUND FORMS OF ARTHROMYCES RAMOSUS PEROXIDASE AT DIFFERENT PH VALUES. PERTURBATIONS OF ACTIVE SITE RESIDUES AND THEIR IMPLICATION IN ENZYME CATALYSIS | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, CALCIUM ION, IODIDE ION, ... | | Authors: | Fukuyama, K, Kunishima, N, Amada, F. | | Deposit date: | 1995-04-25 | | Release date: | 1996-01-29 | | Last modified: | 2020-07-29 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Crystal structures of cyanide- and triiodide-bound forms of Arthromyces ramosus peroxidase at different pH values. Perturbations of active site residues and their implication in enzyme catalysis.
J.Biol.Chem., 270, 1995
|
|
1ARY
 
 | | CRYSTAL STRUCTURES OF CYANIDE-AND TRIIODIDE-BOUND FORMS OF ARTHROMYCES RAMOSUS PEROXIDASE AT DIFFERENT PH VALUES. PERTURBATIONS OF ACTIVE SITE RESIDUES AND THEIR IMPLICATION IN ENZYME CATALYSIS | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, CALCIUM ION, IODIDE ION, ... | | Authors: | Fukuyama, K, Kunishima, N, Amada, F. | | Deposit date: | 1995-04-25 | | Release date: | 1996-01-29 | | Last modified: | 2020-07-29 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Crystal structures of cyanide- and triiodide-bound forms of Arthromyces ramosus peroxidase at different pH values. Perturbations of active site residues and their implication in enzyme catalysis.
J.Biol.Chem., 270, 1995
|
|
6DL7
 
 | | Human mitochondrial ClpP in complex with ONC201 (TIC10) | | Descriptor: | 7-benzyl-4-[(2-methylphenyl)methyl]-6,7,8,9-tetrahydroimidazo[1,2-a]pyrido[3,4-e]pyrimidin-5(4H)-one, ATP-dependent Clp protease proteolytic subunit, mitochondrial | | Authors: | Halgas, O, Zarabi, S.F, Schimmer, A, Pai, E.F. | | Deposit date: | 2018-05-31 | | Release date: | 2019-05-08 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Mitochondrial ClpP-Mediated Proteolysis Induces Selective Cancer Cell Lethality.
Cancer Cell, 35, 2019
|
|
1LGY
 
 | | LIPASE II FROM RHIZOPUS NIVEUS | | Descriptor: | TRIACYLGLYCEROL LIPASE | | Authors: | Kohno, M, Funatsu, J, Mikami, B, Kugimiya, W, Matsuo, T, Morita, Y. | | Deposit date: | 1996-05-23 | | Release date: | 1996-12-23 | | Last modified: | 2024-06-05 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | The crystal structure of lipase II from Rhizopus niveus at 2.2 A resolution.
J.Biochem.(Tokyo), 120, 1996
|
|
1BDM
 
 | |
6AIT
 
 | | Crystal structure of E. coli BepA | | Descriptor: | 2-AMINO-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, Beta-barrel assembly-enhancing protease, ZINC ION | | Authors: | Umar, M.S.M, Tanaka, Y, Kamikubo, H, Tsukazaki, T. | | Deposit date: | 2018-08-24 | | Release date: | 2018-12-26 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (2.598 Å) | | Cite: | Structural Basis for the Function of the beta-Barrel Assembly-Enhancing Protease BepA.
J. Mol. Biol., 431, 2019
|
|
3WDP
 
 | | Structural analysis of a beta-glucosidase mutant derived from a hyperthermophilic tetrameric structure | | Descriptor: | Beta-glucosidase, GLYCEROL, PHOSPHATE ION | | Authors: | Nakabayashi, M, Kataoka, M, Ishikawa, K. | | Deposit date: | 2013-06-19 | | Release date: | 2014-03-12 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Structural analysis of beta-glucosidase mutants derived from a hyperthermophilic tetrameric structure.
Acta Crystallogr.,Sect.D, 70, 2014
|
|
1IS2
 
 | |
8SS0
 
 | | Human sterol 14 alpha-demethylase (CYP51) in complex with the reaction intermediate 14 alpha-aldehyde dihydrolanosterol | | Descriptor: | 3beta-hydroxy-10alpha,13alpha-lanosta-8,24-dien-30-al, Lanosterol 14-alpha demethylase, PROTOPORPHYRIN IX CONTAINING FE | | Authors: | Hargrove, T.Y, Wawrzak, Z, Guengerich, F.P, Lepesheva, G.I. | | Deposit date: | 2023-05-08 | | Release date: | 2024-01-17 | | Last modified: | 2024-03-06 | | Method: | X-RAY DIFFRACTION (2.25 Å) | | Cite: | Oxygen-18 Labeling Reveals a Mixed Fe-O Mechanism in the Last Step of Cytochrome P450 51 Sterol 14 alpha-Demethylation.
Angew.Chem.Int.Ed.Engl., 63, 2024
|
|
1ISR
 
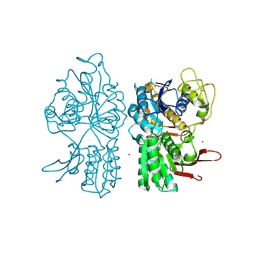 | | Crystal Structure of Metabotropic Glutamate Receptor Subtype 1 Complexed with Glutamate and Gadolinium Ion | | Descriptor: | GADOLINIUM ATOM, GLUTAMIC ACID, Metabotropic Glutamate Receptor subtype 1 | | Authors: | Tsuchiya, D, Kunishima, N, Kamiya, N, Jingami, H, Morikawa, K. | | Deposit date: | 2001-12-21 | | Release date: | 2002-03-13 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (4 Å) | | Cite: | Structural views of the ligand-binding cores of a metabotropic glutamate receptor complexed with an antagonist and both glutamate and Gd3+.
Proc.Natl.Acad.Sci.USA, 99, 2002
|
|
1ISS
 
 | | Crystal Structure of Metabotropic Glutamate Receptor Subtype 1 Complexed with an antagonist | | Descriptor: | (S)-(ALPHA)-METHYL-4-CARBOXYPHENYLGLYCINE, Metabotropic Glutamate Receptor subtype 1 | | Authors: | Tsuchiya, D, Kunishima, N, Kamiya, N, Jingami, H, Morikawa, K. | | Deposit date: | 2001-12-21 | | Release date: | 2002-03-13 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (3.3 Å) | | Cite: | Structural views of the ligand-binding cores of a metabotropic glutamate receptor complexed with an antagonist and both glutamate and Gd3+.
Proc.Natl.Acad.Sci.USA, 99, 2002
|
|
4NI9
 
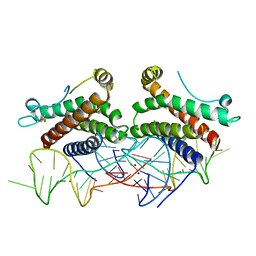 | | Crystal structure of human interleukin 6 in complex with a modified nucleotide aptamer (SOMAMER SL1025), FORM 2 | | Descriptor: | Interleukin-6, SODIUM ION, SOMAmer SL1025 | | Authors: | Davies, D, Edwards, T, Gelinas, A, Jarvis, T, Clifton, M.C. | | Deposit date: | 2013-11-05 | | Release date: | 2014-01-22 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (2.55 Å) | | Cite: | Crystal structure of interleukin-6 in complex with a modified nucleic Acid ligand.
J.Biol.Chem., 289, 2014
|
|
4NI7
 
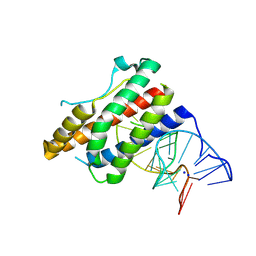 | | Crystal structure of human interleukin 6 in complex with a modified nucleotide aptamer (SOMAMER SL1025) | | Descriptor: | Interleukin-6, SODIUM ION, SOMAmer SL1025 | | Authors: | Davies, D, Edwards, T, Gelinas, A, Jarvis, T, Clifton, M.C. | | Deposit date: | 2013-11-05 | | Release date: | 2014-01-22 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Crystal structure of interleukin-6 in complex with a modified nucleic Acid ligand.
J.Biol.Chem., 289, 2014
|
|
2LWI
 
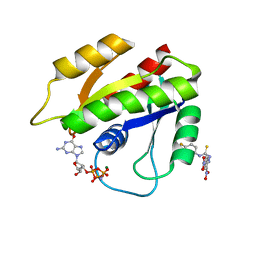 | | Solution structure of H-RasT35S mutant protein in complex with Kobe2601 | | Descriptor: | 2-(2,4-dinitrophenyl)-N-(4-fluorophenyl)hydrazinecarbothioamide, GTPase HRas, MAGNESIUM ION, ... | | Authors: | Araki, M, Tamura, A, Shima, F, Kataoka, T. | | Deposit date: | 2012-08-01 | | Release date: | 2013-05-22 | | Last modified: | 2024-05-15 | | Method: | SOLUTION NMR | | Cite: | In silico discovery of small-molecule Ras inhibitors that display antitumor activity by blocking the Ras-effector interaction.
Proc.Natl.Acad.Sci.USA, 110, 2013
|
|
8D89
 
 | | Crystal structure of a novel GH5 enzyme retrieved from capybara gut metagenome | | Descriptor: | 2-(2-{2-[2-(2-METHOXY-ETHOXY)-ETHOXY]-ETHOXY}-ETHOXY)-ETHANOL, ACETATE ION, CHLORIDE ION, ... | | Authors: | Martins, M.P, Morais, M.A.B, Murakami, M.T. | | Deposit date: | 2022-06-08 | | Release date: | 2022-11-02 | | Last modified: | 2024-05-22 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Glycoside hydrolase subfamily GH5_57 features a highly redesigned catalytic interface to process complex hetero-beta-mannans.
Acta Crystallogr D Struct Biol, 78, 2022
|
|
3FI3
 
 | | Crystal structure of JNK3 with indazole inhibitor, SR-3737 | | Descriptor: | 1,2-ETHANEDIOL, 3-{5-[(2-fluorophenyl)amino]-1H-indazol-1-yl}-N-(3,4,5-trimethoxyphenyl)benzamide, Mitogen-activated protein kinase 10 | | Authors: | Habel, J.E, Duckett, D, LoGrasso, P. | | Deposit date: | 2008-12-10 | | Release date: | 2009-03-03 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Structure-activity relationships and X-ray structures describing the selectivity of aminopyrazole inhibitors for c-Jun N-terminal kinase 3 (JNK3) over p38.
J.Biol.Chem., 284, 2009
|
|
3FI2
 
 | | Crystal structure of JNK3 with amino-pyrazole inhibitor, SR-3451 | | Descriptor: | 1,2-ETHANEDIOL, 3-{4-[(phenylcarbamoyl)amino]-1H-pyrazol-1-yl}-N-(3,4,5-trimethoxyphenyl)benzamide, Mitogen-activated protein kinase 10 | | Authors: | Habel, J.E. | | Deposit date: | 2008-12-10 | | Release date: | 2009-03-03 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (2.28 Å) | | Cite: | Structure-activity relationships and X-ray structures describing the selectivity of aminopyrazole inhibitors for c-Jun N-terminal kinase 3 (JNK3) over p38.
J.Biol.Chem., 284, 2009
|
|
4QPZ
 
 | | Crystal structure of the formolase FLS_v2 in space group P 21 | | Descriptor: | Formolase, MAGNESIUM ION, THIAMINE DIPHOSPHATE | | Authors: | Shen, B.W, Siegel, J.B, Stoddard, B.L, Baker, D. | | Deposit date: | 2014-06-25 | | Release date: | 2015-03-11 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (3 Å) | | Cite: | Computational protein design enables a novel one-carbon assimilation pathway.
Proc.Natl.Acad.Sci.USA, 112, 2015
|
|
4R71
 
 | | Structure of the Qbeta holoenzyme complex in the P1211 crystal form | | Descriptor: | 30S ribosomal protein S1, Elongation factor Ts, Elongation factor Tu, ... | | Authors: | Gytz, H, Seweryn, P, Kutlubaeva, Z, Chetverin, A.B, Brodersen, D.E, Knudsen, C.R. | | Deposit date: | 2014-08-26 | | Release date: | 2015-09-23 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (3.21 Å) | | Cite: | Structural basis for RNA-genome recognition during bacteriophage Q beta replication.
Nucleic Acids Res., 43, 2015
|
|
1C8N
 
 | | TOBACCO NECROSIS VIRUS | | Descriptor: | CALCIUM ION, COAT PROTEIN | | Authors: | Oda, Y, Fukuyama, K. | | Deposit date: | 2000-05-20 | | Release date: | 2000-08-30 | | Last modified: | 2023-08-09 | | Method: | X-RAY DIFFRACTION (2.25 Å) | | Cite: | Crystal structure of tobacco necrosis virus at 2.25 A resolution.
J.Mol.Biol., 300, 2000
|
|
4R06
 
 | | Crystal Structure of SR2067 bound to PPARgamma | | Descriptor: | 1-(naphthalen-1-ylsulfonyl)-N-[(1S)-1-phenylpropyl]-1H-indole-5-carboxamide, Peroxisome proliferator-activated receptor gamma, SULFATE ION | | Authors: | Marrewijk, L, Kamenecka, T, Griffin, P.R, Bruning, J.B. | | Deposit date: | 2014-07-30 | | Release date: | 2016-01-27 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (2.22 Å) | | Cite: | SR2067 Reveals a Unique Kinetic and Structural Signature for PPAR gamma Partial Agonism.
Acs Chem.Biol., 11, 2016
|
|
