6O36
 
 | |
6O46
 
 | |
6U8V
 
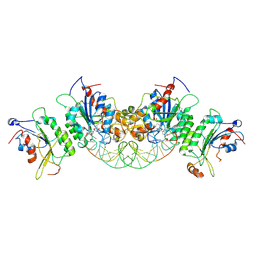 | | Crystal structure of DNMT3B-DNMT3L in complex with CpGpT DNA | | Descriptor: | CpGpT DNA (25-MER), DNA (cytosine-5)-methyltransferase 3-like, DNA (cytosine-5)-methyltransferase 3B, ... | | Authors: | Gao, L, Zhang, Z.M, Song, J. | | Deposit date: | 2019-09-06 | | Release date: | 2020-06-10 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (3 Å) | | Cite: | Comprehensive structure-function characterization of DNMT3B and DNMT3A reveals distinctive de novo DNA methylation mechanisms.
Nat Commun, 11, 2020
|
|
6U8W
 
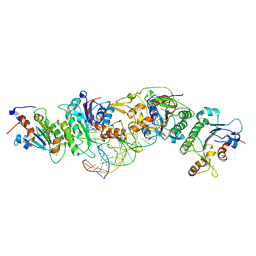 | | Crystal structure of DNMT3B(K777A)-DNMT3L in complex with CpGpT DNA | | Descriptor: | CpGpT DNA (25-MER), DNA (cytosine-5)-methyltransferase 3-like, DNA (cytosine-5)-methyltransferase 3B, ... | | Authors: | Gao, L, Zhang, Z.M, Song, J. | | Deposit date: | 2019-09-06 | | Release date: | 2020-06-10 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (2.94891548 Å) | | Cite: | Comprehensive structure-function characterization of DNMT3B and DNMT3A reveals distinctive de novo DNA methylation mechanisms.
Nat Commun, 11, 2020
|
|
6UX2
 
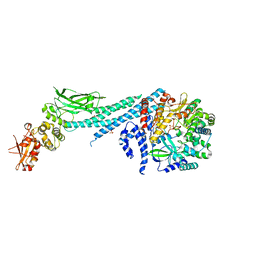 | | Crystal structure of ZIKV RdRp in complex with STAT2 | | Descriptor: | Nonstructural Protein 5, SULFATE ION, Signal transducer and activator of transcription 2, ... | | Authors: | Wang, B, Song, J. | | Deposit date: | 2019-11-06 | | Release date: | 2020-07-08 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (3.01 Å) | | Cite: | Structural basis for STAT2 suppression by flavivirus NS5.
Nat.Struct.Mol.Biol., 27, 2020
|
|
6W7F
 
 | | Structure of EED bound to inhibitor 5285 | | Descriptor: | 8-(6-cyclopropylpyridin-3-yl)-N-[(5-fluoro-2,3-dihydro-1-benzofuran-4-yl)methyl]-1-(methylsulfonyl)imidazo[1,5-c]pyrimidin-5-amine, GLYCEROL, Polycomb protein EED | | Authors: | Petrunak, E.M, Stuckey, J.A. | | Deposit date: | 2020-03-19 | | Release date: | 2020-07-08 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | EEDi-5285: An Exceptionally Potent, Efficacious, and Orally Active Small-Molecule Inhibitor of Embryonic Ectoderm Development.
J.Med.Chem., 63, 2020
|
|
6W7G
 
 | | Structure of EED bound to inhibitor 1056 | | Descriptor: | 8-(2,6-dimethylpyridin-3-yl)-N-[(5-fluoro-2,3-dihydro-1-benzofuran-4-yl)methyl]-1-(methylsulfonyl)imidazo[1,5-c]pyrimidin-5-amine, FORMIC ACID, Polycomb protein EED, ... | | Authors: | Petrunak, E.M, Stuckey, J.A. | | Deposit date: | 2020-03-19 | | Release date: | 2020-07-08 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (1.85 Å) | | Cite: | EEDi-5285: An Exceptionally Potent, Efficacious, and Orally Active Small-Molecule Inhibitor of Embryonic Ectoderm Development.
J.Med.Chem., 63, 2020
|
|
6WCZ
 
 | | CryoEM structure of full-length ZIKV NS5-hSTAT2 complex | | Descriptor: | Non-structural protein 5, Signal transducer and activator of transcription 2, ZINC ION | | Authors: | Boxiao, W, Stephanie, T, Kang, Z, Maria, T.S, Jian, F, Jiuwei, L, Linfeng, G, Wendan, R, Yanxiang, C, Ethan, C.V, HeaJin, H, Matthew, J.E, Sean, E.O, Adolfo, G.S, Hong, Z, Rong, H, Jikui, S. | | Deposit date: | 2020-03-31 | | Release date: | 2020-07-08 | | Last modified: | 2024-05-29 | | Method: | ELECTRON MICROSCOPY (4 Å) | | Cite: | Structural basis for STAT2 suppression by flavivirus NS5.
Nat.Struct.Mol.Biol., 27, 2020
|
|
3D9Y
 
 | |
8DNT
 
 | | SARS-CoV-2 specific T cell receptor | | Descriptor: | Beta-2-microglobulin, MHC class I antigen alpha chain, Nucleoprotein, ... | | Authors: | Gallagher, D.T, Wu, D, Gowthaman, R, Pierce, B.G, Mariuzza, R.A, Weng, N.P. | | Deposit date: | 2022-07-11 | | Release date: | 2023-07-19 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (3.18 Å) | | Cite: | SARS-CoV-2 infection establishes a stable and age-independent CD8 + T cell response against a dominant nucleocapsid epitope using restricted T cell receptors.
Nat Commun, 14, 2023
|
|
6MLT
 
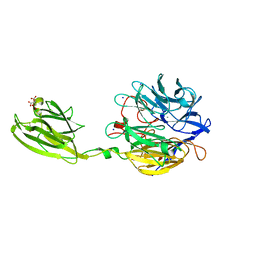 | | Crystal structure of the V. cholerae biofilm matrix protein Bap1 | | Descriptor: | CALCIUM ION, CITRATE ANION, GLYCEROL, ... | | Authors: | Kaus, K, Biester, A, Chupp, E, Lu, K, Vidsudharomn, C, Olson, R. | | Deposit date: | 2018-09-28 | | Release date: | 2019-08-28 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | The 1.9 angstrom crystal structure of the extracellular matrix protein Bap1 fromVibrio choleraeprovides insights into bacterial biofilm adhesion.
J.Biol.Chem., 294, 2019
|
|
3DAV
 
 | |
7MSD
 
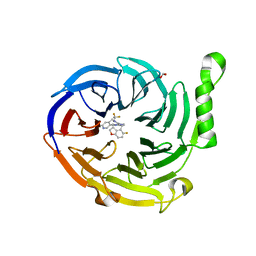 | | Structure of EED bound to EEDi-6068 | | Descriptor: | (9aP,12aR)-4-(2,2-difluoropropyl)-12-{[(5-fluoro-2,3-dihydro-1-benzofuran-4-yl)methyl]amino}-7-(trifluoromethyl)-4,5-dihydro-3H-2,4,8,11,12a-pentaazabenzo[4,5]cycloocta[1,2,3-cd]inden-3-one, FORMIC ACID, Polycomb protein EED | | Authors: | Petrunak, E, Stuckey, J. | | Deposit date: | 2021-05-11 | | Release date: | 2021-10-20 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Discovery of EEDi-5273 as an Exceptionally Potent and Orally Efficacious EED Inhibitor Capable of Achieving Complete and Persistent Tumor Regression.
J.Med.Chem., 64, 2021
|
|
7MSB
 
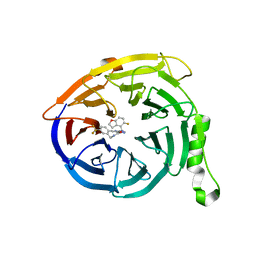 | | Structure of EED bound to EEDi-4259 | | Descriptor: | (9aM,12aS)-12-{[(5-fluoro-1-benzofuran-4-yl)methyl]amino}-7-(trifluoromethyl)-4,5-dihydro-3H-2,4,11,12a-tetraazabenzo[4,5]cycloocta[1,2,3-cd]inden-3-one, Polycomb protein EED | | Authors: | Petrunak, E, Stuckey, J. | | Deposit date: | 2021-05-11 | | Release date: | 2021-10-20 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Discovery of EEDi-5273 as an Exceptionally Potent and Orally Efficacious EED Inhibitor Capable of Achieving Complete and Persistent Tumor Regression.
J.Med.Chem., 64, 2021
|
|
6W8V
 
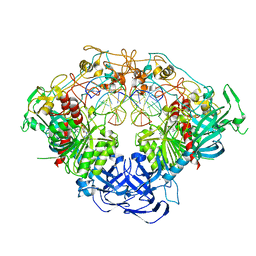 | | Crystal structure of mouse DNMT1 in complex with ACG DNA | | Descriptor: | ACG DNA (5'-D(*AP*CP*TP*TP*AP*(C49)P*GP*GP*AP*AP*GP*G)-3'), ACG DNA (5'-D(*CP*CP*TP*TP*CP*(5CM)P*GP*TP*AP*AP*GP*T)-3'), DNA (cytosine-5)-methyltransferase 1, ... | | Authors: | Anteneh, H, Song, J. | | Deposit date: | 2020-03-21 | | Release date: | 2020-07-08 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (3.12 Å) | | Cite: | DNMT1 activity, base flipping mechanism and genome-wide DNA methylation are regulated by the DNA sequence context
Nat Commun, 2020
|
|
6W8W
 
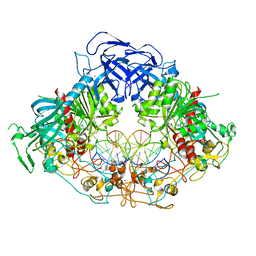 | | Crystal structure of mouse DNMT1 in complex with CCG DNA | | Descriptor: | CCG DNA (5'-D(*AP*CP*TP*TP*AP*(5CM)P*GP*GP*AP*AP*GP*G)-3'), CCG DNA (5'-D(*CP*CP*TP*TP*CP*(C49)P*GP*TP*AP*AP*GP*T)-3'), DNA (cytosine-5)-methyltransferase 1, ... | | Authors: | Anteneh, H, Song, J. | | Deposit date: | 2020-03-21 | | Release date: | 2020-07-08 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (3 Å) | | Cite: | DNMT1 activity, base flipping mechanism and genome-wide DNA methylation are regulated by the DNA sequence context
Nat Commun, 2020
|
|
7L4C
 
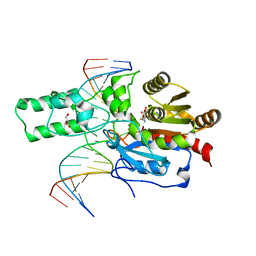 | | Crystal structure of the DRM2-CTT DNA complex | | Descriptor: | DNA (5'-D(*AP*TP*TP*AP*TP*TP*AP*AP*TP*(C49)P*TP*TP*AP*AP*TP*TP*TP*A)-3'), DNA (5'-D(*TP*AP*AP*AP*TP*TP*AP*AP*GP*AP*TP*TP*AP*AP*TP*AP*AP*T)-3'), DNA (cytosine-5)-methyltransferase DRM2, ... | | Authors: | Fang, J, Song, J. | | Deposit date: | 2020-12-18 | | Release date: | 2021-08-04 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (2.11 Å) | | Cite: | Substrate deformation regulates DRM2-mediated DNA methylation in plants.
Sci Adv, 7, 2021
|
|
7L4K
 
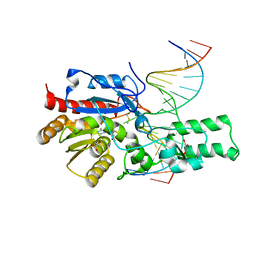 | | Crystal structure of the DRM2-CCG DNA complex | | Descriptor: | DNA (5'-D(*AP*TP*TP*CP*CP*TP*AP*AP*TP*(C49)P*CP*GP*AP*AP*TP*TP*TP*A)-3'), DNA (5'-D(*TP*AP*AP*AP*TP*TP*CP*GP*GP*AP*TP*TP*AP*GP*GP*AP*AP*T)-3'), DNA (cytosine-5)-methyltransferase DRM2, ... | | Authors: | Fang, J, Song, J. | | Deposit date: | 2020-12-19 | | Release date: | 2021-08-04 | | Last modified: | 2024-10-09 | | Method: | X-RAY DIFFRACTION (2.61 Å) | | Cite: | Substrate deformation regulates DRM2-mediated DNA methylation in plants.
Sci Adv, 7, 2021
|
|
7L4H
 
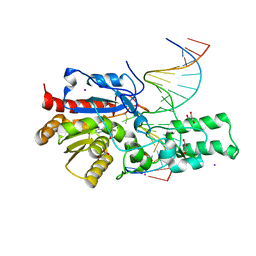 | | Crystal structure of the DRM2-CTG DNA complex | | Descriptor: | DNA (5'-D(*AP*TP*TP*CP*CP*TP*AP*AP*TP*(C49)P*TP*GP*AP*AP*TP*TP*TP*A)-3'), DNA (5'-D(*TP*AP*AP*AP*TP*TP*CP*AP*GP*AP*TP*TP*AP*GP*GP*AP*AP*T)-3'), DNA (cytosine-5)-methyltransferase DRM2, ... | | Authors: | Fang, J, Song, J. | | Deposit date: | 2020-12-19 | | Release date: | 2021-08-04 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (2.56 Å) | | Cite: | Substrate deformation regulates DRM2-mediated DNA methylation in plants.
Sci Adv, 7, 2021
|
|
7L4N
 
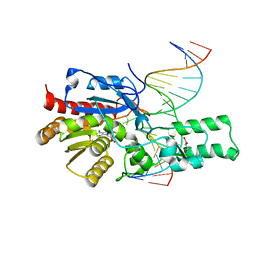 | | Crystal structure of the DRM2 (C397R)-CCG DNA complex | | Descriptor: | DNA (5'-D(*AP*TP*TP*CP*CP*TP*AP*AP*TP*(C49)P*CP*GP*AP*AP*TP*TP*TP*A)-3'), DNA (5'-D(*TP*AP*AP*AP*TP*TP*CP*GP*GP*AP*TP*TP*AP*GP*GP*AP*AP*T)-3'), DNA (cytosine-5)-methyltransferase DRM2, ... | | Authors: | Fang, J, Song, J. | | Deposit date: | 2020-12-19 | | Release date: | 2021-08-04 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (2.247 Å) | | Cite: | Substrate deformation regulates DRM2-mediated DNA methylation in plants.
Sci Adv, 7, 2021
|
|
7L4M
 
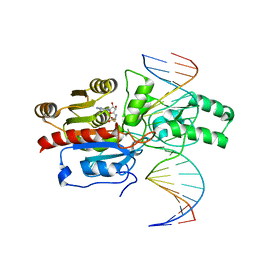 | | Crystal structure of the DRM2-CCT DNA complex | | Descriptor: | DNA (5'-D(*TP*AP*AP*AP*GP*GP*AP*GP*GP*AP*GP*GP*AP*GP*GP*AP*AP*T)-3'), DNA (5'-D(P*AP*TP*TP*CP*CP*TP*CP*CP*TP*(C49)P*CP*TP*CP*CP*TP*TP*TP*A)-3'), DNA (cytosine-5)-methyltransferase DRM2, ... | | Authors: | Fang, J, Song, J. | | Deposit date: | 2020-12-19 | | Release date: | 2021-08-04 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (2.805 Å) | | Cite: | Substrate deformation regulates DRM2-mediated DNA methylation in plants.
Sci Adv, 7, 2021
|
|
7L4F
 
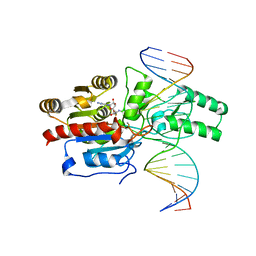 | | Crystal structure of the DRM2-CAT DNA complex | | Descriptor: | 2'-DEOXYADENOSINE-5'-MONOPHOSPHATE, DNA (5'-D(*AP*TP*TP*CP*CP*TP*CP*CP*TP*(C49)P*AP*TP*CP*CP*TP*TP*TP*A)-3'), DNA (5'-D(*TP*AP*AP*AP*GP*GP*AP*TP*GP*AP*GP*GP*AP*GP*GP*AP*AP*T)-3'), ... | | Authors: | Fang, J, Song, J. | | Deposit date: | 2020-12-19 | | Release date: | 2021-08-04 | | Last modified: | 2024-05-29 | | Method: | X-RAY DIFFRACTION (2.55 Å) | | Cite: | Substrate deformation regulates DRM2-mediated DNA methylation in plants.
Sci Adv, 7, 2021
|
|
8GJN
 
 | | 17B10 fab in complex with up-RBD of SARS-CoV-2 Spike G614 trimer | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, Heavy chain of 17B10 Fab, Light chain of 17B10 Fab, ... | | Authors: | Kwon, H.J, Zhang, J, Kosikova, M, Tang, W.C, Rodriguez, U.O, Peng, H.Q, Meseda, C.A, Pedro, C.L, Schmeisser, F, Lu, J.M, Zhou, B, Davis, C.T, Wentworth, D.E, Chen, W.H, Shriver, M.C, Pasetti, M.F, Weir, J.P, Chen, B, Xie, H. | | Deposit date: | 2023-03-16 | | Release date: | 2023-04-05 | | Last modified: | 2024-10-23 | | Method: | ELECTRON MICROSCOPY (3.6 Å) | | Cite: | Distinct in vitro and in vivo neutralization profiles of monoclonal antibodies elicited by the receptor binding domain of the ancestral SARS-CoV-2.
J Med Virol, 95, 2023
|
|
7DAC
 
 | | Human RIPK3 amyloid fibril revealed by solid-state NMR | | Descriptor: | Receptor-interacting serine/threonine-protein kinase 3 | | Authors: | Wu, X.L, Zhang, J, Dong, X.Q, Liu, J, Li, B, Hu, H, Wang, J, Wang, H.Y, Lu, J.X. | | Deposit date: | 2020-10-16 | | Release date: | 2021-04-28 | | Last modified: | 2024-05-01 | | Method: | SOLID-STATE NMR | | Cite: | The structure of a minimum amyloid fibril core formed by necroptosis-mediating RHIM of human RIPK3.
Proc.Natl.Acad.Sci.USA, 118, 2021
|
|
7XLB
 
 | |
