4QTC
 
 | | Structure of human haspin (GSG2) in complex with SCH772984 revealing the first type-I binding mode | | Descriptor: | (3R)-1-(2-oxo-2-{4-[4-(pyrimidin-2-yl)phenyl]piperazin-1-yl}ethyl)-N-[3-(pyridin-4-yl)-2H-indazol-5-yl]pyrrolidine-3-carboxamide, (4S)-2-METHYL-2,4-PENTANEDIOL, GLYCEROL, ... | | Authors: | Chaikuad, A, von Delft, F, Arrowsmith, C.H, Edwards, A.M, Bountra, C, Knapp, S, Structural Genomics Consortium (SGC) | | Deposit date: | 2014-07-07 | | Release date: | 2014-07-23 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (1.4 Å) | | Cite: | A unique inhibitor binding site in ERK1/2 is associated with slow binding kinetics.
Nat.Chem.Biol., 10, 2014
|
|
5K47
 
 | | CryoEM structure of the human Polycystin-2/PKD2 TRP channel | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Polycystin-2 | | Authors: | Pike, A.C.W, Grieben, M, Shintre, C.A, Tessitore, A, Shrestha, L, Mukhopadhyay, S, Mahajan, P, Chalk, R, Burgess-Brown, N.A, Huiskonen, J.T, Arrowsmith, C.H, Edwards, A.M, Bountra, C, Carpenter, E.P, Structural Genomics Consortium (SGC) | | Deposit date: | 2016-05-20 | | Release date: | 2016-08-24 | | Last modified: | 2020-10-21 | | Method: | ELECTRON MICROSCOPY (4.2 Å) | | Cite: | Structure of the polycystic kidney disease TRP channel Polycystin-2 (PC2).
Nat. Struct. Mol. Biol., 24, 2017
|
|
5KCH
 
 | | SETDB1 in complex with an early stage, low affinity fragment candidate modelled at reduced occupancy into weak electron density | | Descriptor: | 4-methoxy-N-[(pyridin-2-yl)methyl]aniline, DIMETHYL SULFOXIDE, Histone-lysine N-methyltransferase SETDB1, ... | | Authors: | Tempel, W, Harding, R.J, Mader, P, Dobrovetsky, E, Walker, J.R, Brown, P.J, Schapira, M, Collins, P, Pearce, N, Brandao-Neto, J, Douangamath, A, von Delft, F, Bountra, C, Arrowsmith, C.H, Edwards, A.M, Santhakumar, V, Structural Genomics Consortium (SGC) | | Deposit date: | 2016-06-06 | | Release date: | 2016-07-27 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | SETDB1 in complex with an early stage, low affinity fragment candidate modelled at reduced occupancy
To Be Published
|
|
4A63
 
 | | Crystal structure of the p73-ASPP2 complex at 2.6A resolution | | Descriptor: | ACETATE ION, APOPTOSIS STIMULATING OF P53 PROTEIN 2, TUMOUR PROTEIN 73, ... | | Authors: | Canning, P, Sharpe, T, Krojer, T, Savitsky, P, Cooper, C.D.O, Salah, E, Keates, T, Muniz, J, Vollmar, M, von Delft, F, Weigelt, J, Arrowsmith, C, Bountra, C, Edwards, A, Bullock, A.N. | | Deposit date: | 2011-10-31 | | Release date: | 2011-12-21 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (2.27 Å) | | Cite: | Structural Basis for Aspp2 Recognition by the Tumor Suppressor P73.
J.Mol.Biol., 423, 2012
|
|
5KCO
 
 | | SETDB1 in complex with an early stage, low affinity fragment candidate modelled at reduced occupancy | | Descriptor: | DIMETHYL SULFOXIDE, Histone-lysine N-methyltransferase SETDB1, SULFATE ION, ... | | Authors: | Tempel, W, Harding, R.J, Mader, P, Dobrovetsky, E, Walker, J.R, Brown, P.J, Schapira, M, Collins, P, Pearce, N, Brandao-Neto, J, Douangamath, A, von Delft, F, Bountra, C, Arrowsmith, C.H, Edwards, A.M, Santhakumar, V, Structural Genomics Consortium (SGC) | | Deposit date: | 2016-06-06 | | Release date: | 2016-07-27 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (1.47 Å) | | Cite: | SETDB1 in complex with an early stage, low affinity fragment candidate modelled at reduced occupancy
To Be Published
|
|
5KH9
 
 | | Crystal structure of a low occupancy fragment candidate (5-[(4-Isopropylphenyl)amino]-6-methyl-1,2,4-triazin-3(2H)-one) bound in the ubiquitin binding pocket of the HDAC6 zinc-finger domain | | Descriptor: | 6-methyl-5-[(4-propan-2-ylphenyl)amino]-2~{H}-1,2,4-triazin-3-one, FORMIC ACID, Histone deacetylase 6, ... | | Authors: | Harding, R.J, Tempel, W, Ravichandran, M, Collins, P, Pearce, N, Brandao-Neto, J, Douangamath, A, Schapira, M, Bountra, C, Edwards, A.M, von Delft, F, Santhakumar, V, Arrowsmith, C.M, Structural Genomics Consortium (SGC) | | Deposit date: | 2016-06-14 | | Release date: | 2016-07-27 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (1.07 Å) | | Cite: | Small Molecule Antagonists of the Interaction between the Histone Deacetylase 6 Zinc-Finger Domain and Ubiquitin.
J. Med. Chem., 60, 2017
|
|
5KH7
 
 | | Crystal structure of fragment (3-[6-Oxo-3-(3-pyridinyl)-1(6H)-pyridazinyl]propanoic acid) bound in the ubiquitin binding pocket of the HDAC6 zinc-finger domain | | Descriptor: | 3-(6-oxidanylidene-3-pyridin-3-yl-pyridazin-1-yl)propanoic acid, Histone deacetylase 6, UNKNOWN ATOM OR ION, ... | | Authors: | Harding, R.J, Walker, J, Ravichandran, M, Ferreira de Freitas, R, Schapira, M, Bountra, C, Edwards, A.M, Santhakumar, V, Arrowsmith, C.M, Structural Genomics Consortium (SGC) | | Deposit date: | 2016-06-14 | | Release date: | 2016-07-27 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Small Molecule Antagonists of the Interaction between the Histone Deacetylase 6 Zinc-Finger Domain and Ubiquitin.
J. Med. Chem., 60, 2017
|
|
5J1W
 
 | | Crystal structure of human CLK1 in complex with pyrido[3,4-g]quinazoline derivative ZW31 (compound 14) | | Descriptor: | Dual specificity protein kinase CLK1, GLYCEROL, PHOSPHATE ION, ... | | Authors: | Chaikuad, A, Esvan, Y.J, Zeinyeh, W, Boibessot, T, Nauton, L, Thery, V, Loaec, N, Meijer, L, Giraud, F, Moreau, P, Anizon, F, von Delft, F, Bountra, C, Arrowsmith, C.H, Edwards, A.M, Knapp, S, Structural Genomics Consortium (SGC) | | Deposit date: | 2016-03-29 | | Release date: | 2016-05-04 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (2.42 Å) | | Cite: | Discovery of pyrido[3,4-g]quinazoline derivatives as CMGC family protein kinase inhibitors: Design, synthesis, inhibitory potency and X-ray co-crystal structure.
Eur.J.Med.Chem., 118, 2016
|
|
5J1V
 
 | | Crystal structure of human CLK1 in complex with pyrido[3,4-g]quinazoline derivative ZW29 (compound 13) | | Descriptor: | Dual specificity protein kinase CLK1, GLYCEROL, pyrido[3,4-g]quinazoline-2,10-diamine | | Authors: | Chaikuad, A, Esvan, Y.J, Zeinyeh, W, Boibessot, T, Nauton, L, Thery, V, Loaec, N, Meijer, L, Giraud, F, Moreau, P, Anizon, F, von Delft, F, Bountra, C, Arrowsmith, C.H, Edwards, A.M, Knapp, S, Structural Genomics Consortium (SGC) | | Deposit date: | 2016-03-29 | | Release date: | 2016-05-04 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (2.52 Å) | | Cite: | Discovery of pyrido[3,4-g]quinazoline derivatives as CMGC family protein kinase inhibitors: Design, synthesis, inhibitory potency and X-ray co-crystal structure.
Eur.J.Med.Chem., 118, 2016
|
|
4B9D
 
 | | Crystal Structure of Human NIMA-related Kinase 1 (NEK1) with inhibitor. | | Descriptor: | 1,2-ETHANEDIOL, CHLORIDE ION, SERINE/THREONINE-PROTEIN KINASE NEK1, ... | | Authors: | Elkins, J.M, Hanchuk, T.D.M, Lovato, D.V, Basei, F.L, Meirelles, G.V, Kobarg, J, Szklarz, M, Vollmar, M, Mahajan, P, Rellos, P, Zhang, Y, Krojer, T, Pike, A.C.W, Canning, P, von Delft, F, Raynor, J, Bountra, C, Arrowsmith, C, Edwards, A, Knapp, S. | | Deposit date: | 2012-09-04 | | Release date: | 2012-09-19 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | NEK1 kinase domain structure and its dynamic protein interactome after exposure to Cisplatin.
Sci Rep, 7, 2017
|
|
4APC
 
 | | Crystal Structure of Human NIMA-related Kinase 1 (NEK1) | | Descriptor: | CHLORIDE ION, SERINE/THREONINE-PROTEIN KINASE NEK1 | | Authors: | Elkins, J.M, Hanchuk, T.D.M, Lovato, D.V, Basei, F.L, Meirelles, G.V, Kobarg, J, Szklarz, M, Vollmar, M, Mahajan, P, Rellos, P, Zhang, Y, Krojer, T, Pike, A.C.W, Bountra, C, Arrowsmith, C, Edwards, A, Knapp, S. | | Deposit date: | 2012-04-02 | | Release date: | 2012-04-25 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | NEK1 kinase domain structure and its dynamic protein interactome after exposure to Cisplatin.
Sci Rep, 7, 2017
|
|
5LAR
 
 | | Crystal structure of p38 alpha MAPK14 in complex with VPC00628 | | Descriptor: | 5-azanyl-~{N}-[[4-[[(2~{S})-1-azanyl-4-cyclohexyl-1-oxidanylidene-butan-2-yl]carbamoyl]phenyl]methyl]-1-phenyl-pyrazole-4-carboxamide, Mitogen-activated protein kinase 14 | | Authors: | Chaikuad, A, Petersen, L.K, von Delft, F, Arrowsmith, C.H, Edwards, A.M, Bountra, C, Knapp, S, Structural Genomics Consortium (SGC) | | Deposit date: | 2016-06-14 | | Release date: | 2016-07-06 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | Novel p38alpha MAP kinase inhibitors identified from yoctoReactor DNA-encoded small molecule library
Medchemcomm, 7, 2016
|
|
5KO9
 
 | | Crystal Structure of the SRAP Domain of Human HMCES Protein | | Descriptor: | Embryonic stem cell-specific 5-hydroxymethylcytosine-binding protein, UNKNOWN ATOM OR ION | | Authors: | Halabelian, L, Li, Y, Bountra, C, Edwards, A.M, Arrowsmith, C.H, Structural Genomics Consortium (SGC) | | Deposit date: | 2016-06-29 | | Release date: | 2016-08-24 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | Structural basis of HMCES interactions with abasic DNA and multivalent substrate recognition.
Nat.Struct.Mol.Biol., 26, 2019
|
|
4ABN
 
 | | Crystal structure of full length mouse Strap (TTC5) | | Descriptor: | 1,2-ETHANEDIOL, TETRATRICOPEPTIDE REPEAT PROTEIN 5 | | Authors: | Pike, A.C.W, Bullock, A.N, Kleinekofort, W, Zimmermann, T, Burgess-Brown, N, Sharpe, T.D, Thangaratnarajah, C, Keates, T, Ugochukwu, E, Bunkoczi, G, Uppenberg, J, von Delft, F, Arrowsmith, C.H, Weigelt, J, Edwards, A, Bountra, C, La Thangue, N.B, Knapp, S. | | Deposit date: | 2011-12-09 | | Release date: | 2012-01-25 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (2.05 Å) | | Cite: | The P53 Cofactor Strap Exhibits an Unexpected Tpr Motif and Oligonucleotide-Binding (Ob)-Fold Structure.
Proc.Natl.Acad.Sci.USA, 109, 2012
|
|
5LB3
 
 | | Crystal structure of human RECQL5 helicase in complex with ADP/Mg. | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, ATP-dependent DNA helicase Q5, MAGNESIUM ION, ... | | Authors: | Newman, J.A, Aitkenhead, H, Savitsky, P, Krojer, T, von Delft, F, Arrowsmith, C.H, Edwards, A.M, Bountra, C, Gileadi, O, Structural Genomics Consortium (SGC) | | Deposit date: | 2016-06-15 | | Release date: | 2016-07-06 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Insights into the RecQ helicase mechanism revealed by the structure of the helicase domain of human RECQL5.
Nucleic Acids Res., 45, 2017
|
|
4B99
 
 | | Crystal Structure of MAPK7 (ERK5) with inhibitor | | Descriptor: | 11-cyclopentyl-2-[[2-methoxy-4-[4-(4-methylpiperazin-1-yl)piperidin-1-yl]carbonyl-phenyl]amino]-5-methyl-pyrimido[4,5-b][1,4]benzodiazepin-6-one, MITOGEN-ACTIVATED PROTEIN KINASE 7 | | Authors: | Elkins, J.M, Wang, J, Vollmar, M, Mahajan, P, Savitsky, P, Deng, X, Gray, N.S, Pike, A.C.W, von Delft, F, Bountra, C, Arrowsmith, C, Edwards, A, Knapp, S. | | Deposit date: | 2012-09-03 | | Release date: | 2012-09-19 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | X-Ray Crystal Structure of Erk5 (Mapk7) in Complex with a Specific Inhibitor.
J.Med.Chem., 56, 2013
|
|
4BKN
 
 | | Human Dihydropyrimidinase-related protein 3 (DPYSL3) | | Descriptor: | DIHYDROPYRIMIDINASE-RELATED PROTEIN 3 | | Authors: | Mathea, S, Elkins, J.M, Alegre-Abarrategui, J, Shrestha, L, Burgess-Brown, N, Puranik, S, Coutandin, D, Bradley, A, Vollmar, M, von Delft, F, Bountra, C, Arrowsmith, C, Edwards, A, Knapp, S. | | Deposit date: | 2013-04-26 | | Release date: | 2013-05-08 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Structure of Dpysl3
To be Published
|
|
6I5I
 
 | | Crystal structure of CLK1 in complexed with furo[3,2-b]pyridine compound 12h | | Descriptor: | 1,2-ETHANEDIOL, 5-(1-methylpyrazol-4-yl)-3-[1-(phenylmethyl)pyrazol-4-yl]furo[3,2-b]pyridine, Dual specificity protein kinase CLK1 | | Authors: | Chaikuad, A, Arrowsmith, C.H, Edwards, A.M, Bountra, C, Paruch, K, Knapp, S, Structural Genomics Consortium (SGC) | | Deposit date: | 2018-11-13 | | Release date: | 2019-01-09 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Furo[3,2-b]pyridine: A Privileged Scaffold for Highly Selective Kinase Inhibitors and Effective Modulators of the Hedgehog Pathway.
Angew. Chem. Int. Ed. Engl., 58, 2019
|
|
6HPX
 
 | | Crystal structure of ENL (MLLT1) in complex with compound 19 | | Descriptor: | 1,2-ETHANEDIOL, Protein ENL, ~{N}-[(3-chlorophenyl)methyl]-1-(2-pyrrolidin-1-ylethyl)benzimidazole-5-carboxamide | | Authors: | Heidenreich, D, Chaikuad, A, Arrowsmith, C.H, Edwards, A.M, Bountra, C, Knapp, S, Structural Genomics Consortium (SGC) | | Deposit date: | 2018-09-22 | | Release date: | 2018-11-28 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Structure-Based Approach toward Identification of Inhibitory Fragments for Eleven-Nineteen-Leukemia Protein (ENL).
J.Med.Chem., 61, 2018
|
|
6HPZ
 
 | | Crystal structure of ENL (MLLT1) in complex with acetyllysine | | Descriptor: | 1,2-ETHANEDIOL, N(6)-ACETYLLYSINE, Protein ENL | | Authors: | Heidenreich, D, Chaikuad, A, Arrowsmith, C.H, Edwards, A.M, Bountra, C, Knapp, S, Structural Genomics Consortium (SGC) | | Deposit date: | 2018-09-22 | | Release date: | 2018-11-28 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Structure-Based Approach toward Identification of Inhibitory Fragments for Eleven-Nineteen-Leukemia Protein (ENL).
J.Med.Chem., 61, 2018
|
|
6TSZ
 
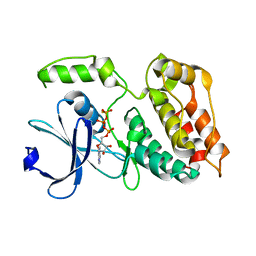 | | The ULK4 Pseudokinase Domain Bound To ATPgammaS | | Descriptor: | PHOSPHOTHIOPHOSPHORIC ACID-ADENYLATE ESTER, Serine/threonine-protein kinase ULK4 | | Authors: | Preuss, F, Chatterjee, D, Mathea, S, Arrowsmith, C.H, Bountra, C, Edwards, A.M, Knapp, S. | | Deposit date: | 2019-12-22 | | Release date: | 2020-01-01 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Nucleotide Binding, Evolutionary Insights, and Interaction Partners of the Pseudokinase Unc-51-like Kinase 4.
Structure, 28, 2020
|
|
6HPY
 
 | | Crystal structure of ENL (MLLT1) in complex with compound 12 | | Descriptor: | 1,2-ETHANEDIOL, 3-[4-[(4-~{tert}-butylphenyl)carbonylamino]phenyl]propanoic acid, Protein ENL, ... | | Authors: | Heidenreich, D, Chaikuad, A, Arrowsmith, C.H, Edwards, A.M, Bountra, C, Knapp, S, Structural Genomics Consortium (SGC) | | Deposit date: | 2018-09-22 | | Release date: | 2018-11-28 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Structure-Based Approach toward Identification of Inhibitory Fragments for Eleven-Nineteen-Leukemia Protein (ENL).
J.Med.Chem., 61, 2018
|
|
6HP9
 
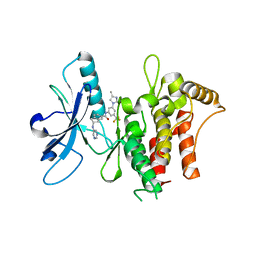 | | Structure of the kinase domain of human DDR1 in complex with a 2-Amino-2,3-Dihydro-1H-Indene-5-Carboxamide-based inhibitor | | Descriptor: | (2~{R})-~{N}-[3-(4-methylimidazol-1-yl)-5-(trifluoromethyl)phenyl]-2-(pyrimidin-5-ylamino)-2,3-dihydro-1~{H}-indene-5-carboxamide, Epithelial discoidin domain-containing receptor 1 | | Authors: | Pinkas, D.M, Fox, A.E, Kupinska, K, Burgess-Brown, N.A, von Delft, F, Arrowsmith, C.H, Edwards, A.M, Bountra, C, Bullock, A.N, Structural Genomics Consortium (SGC) | | Deposit date: | 2018-09-19 | | Release date: | 2019-01-30 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (1.96 Å) | | Cite: | 2-Amino-2,3-dihydro-1H-indene-5-carboxamide-Based Discoidin Domain Receptor 1 (DDR1) Inhibitors: Design, Synthesis, and in Vivo Antipancreatic Cancer Efficacy.
J.Med.Chem., 62, 2019
|
|
6IBD
 
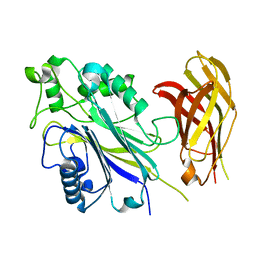 | | The Phosphatase and C2 domains of Human SHIP1 | | Descriptor: | Phosphatidylinositol 3,4,5-trisphosphate 5-phosphatase 1 | | Authors: | Bradshaw, W.J, Williams, E.P, Fernandez-Cid, A, Burgess-Brown, N, von Delft, F, Arrowsmith, C.H, Edwards, A, Bountra, C, Gileadi, O. | | Deposit date: | 2018-11-29 | | Release date: | 2019-01-16 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (1.48 Å) | | Cite: | Regulation of inositol 5-phosphatase activity by the C2 domain of SHIP1 and SHIP2.
Structure, 2024
|
|
6I5L
 
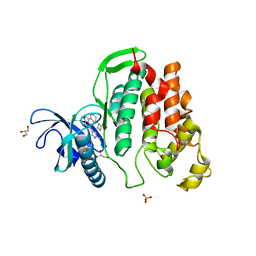 | | Crystal structure of CLK1 in complexed with furo[3,2-b]pyridine compound VN316 (derivative of compound 12h) | | Descriptor: | 3-(3-cyclobutylphenyl)-5-(1-methylpyrazol-4-yl)furo[3,2-b]pyridine, Dual specificity protein kinase CLK1, GLYCEROL, ... | | Authors: | Chaikuad, A, Arrowsmith, C.H, Edwards, A.M, Bountra, C, Paruch, K, Knapp, S, Structural Genomics Consortium (SGC) | | Deposit date: | 2018-11-13 | | Release date: | 2019-01-09 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (2.55 Å) | | Cite: | Furo[3,2-b]pyridine: A Privileged Scaffold for Highly Selective Kinase Inhibitors and Effective Modulators of the Hedgehog Pathway.
Angew. Chem. Int. Ed. Engl., 58, 2019
|
|
