3PTE
 
 | |
1QPE
 
 | |
3EUO
 
 | |
3EUQ
 
 | |
3EUT
 
 | |
3F5O
 
 | | Crystal Structure of hTHEM2(undecan-2-one-CoA) complex | | Descriptor: | CHLORIDE ION, COENZYME A, HEXAETHYLENE GLYCOL, ... | | Authors: | Xu, H, Gong, W. | | Deposit date: | 2008-11-04 | | Release date: | 2009-08-18 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | The mechanisms of human hotdog-fold thioesterase 2 (hTHEM2) substrate recognition and catalysis illuminated by a structure and function based analysis
Biochemistry, 48, 2009
|
|
3QQU
 
 | | Cocrystal structure of unphosphorylated igf with pyrimidine 8 | | Descriptor: | Insulin-like growth factor 1 receptor, N~2~-[3-methoxy-4-(morpholin-4-yl)phenyl]-N~4~-(quinolin-3-yl)pyrimidine-2,4-diamine | | Authors: | Huang, X. | | Deposit date: | 2011-02-16 | | Release date: | 2011-04-20 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (2.9 Å) | | Cite: | Discovery of 2,4-bis-arylamino-1,3-pyrimidines as insulin-like growth factor-1 receptor (IGF-1R) inhibitors.
Bioorg.Med.Chem.Lett., 21, 2011
|
|
3OHI
 
 | | Structure of Giardia fructose-1,6-biphosphate aldolase in complex with 3-hydroxy-2-pyridone | | Descriptor: | ({3-hydroxy-2-oxo-4-[2-(phosphonooxy)ethyl]pyridin-1(2H)-yl}methyl)phosphonic acid, Putative fructose-1,6-bisphosphate aldolase, ZINC ION | | Authors: | Herzberg, O, Galkin, A. | | Deposit date: | 2010-08-17 | | Release date: | 2011-01-19 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Rational design, synthesis and evaluation of first generation inhibitors of the Giardia lamblia fructose-1,6-biphosphate aldolase.
J.Inorg.Biochem., 105, 2010
|
|
3UYS
 
 | | Crystal structure of apo human ck1d | | Descriptor: | Casein kinase I isoform delta, SULFATE ION | | Authors: | Huang, X. | | Deposit date: | 2011-12-06 | | Release date: | 2012-01-11 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Structural basis for the interaction between casein kinase 1 delta and a potent and selective inhibitor.
J.Med.Chem., 55, 2012
|
|
3UYT
 
 | | crystal structure of ck1d with PF670462 from P1 crystal form | | Descriptor: | 4-[1-cyclohexyl-4-(4-fluorophenyl)-1H-imidazol-5-yl]pyrimidin-2-amine, Casein kinase I isoform delta, SULFATE ION | | Authors: | Huang, X. | | Deposit date: | 2011-12-06 | | Release date: | 2012-01-11 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Structural basis for the interaction between casein kinase 1 delta and a potent and selective inhibitor.
J.Med.Chem., 55, 2012
|
|
3UNM
 
 | | Crystal Structure of The Human MDC1 FHA Domain | | Descriptor: | Mediator of DNA damage checkpoint protein 1 | | Authors: | Luo, S, Ye, K. | | Deposit date: | 2011-11-16 | | Release date: | 2012-01-25 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Structural mechanism of the phosphorylation-dependent dimerization of the MDC1 forkhead-associated domain
Nucleic Acids Res., 40, 2012
|
|
3UZP
 
 | | crystal structure of ck1d with PF670462 from P21 crystal form | | Descriptor: | 4-[1-cyclohexyl-4-(4-fluorophenyl)-1H-imidazol-5-yl]pyrimidin-2-amine, Casein kinase I isoform delta | | Authors: | Huang, X. | | Deposit date: | 2011-12-07 | | Release date: | 2012-01-11 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (1.94 Å) | | Cite: | Structural basis for the interaction between casein kinase 1 delta and a potent and selective inhibitor.
J.Med.Chem., 55, 2012
|
|
2FTK
 
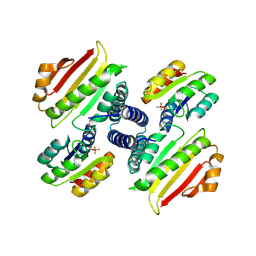 | | berylloflouride Spo0F complex with Spo0B | | Descriptor: | MAGNESIUM ION, Sporulation initiation phosphotransferase B, Sporulation initiation phosphotransferase F | | Authors: | Varughese, K.I. | | Deposit date: | 2006-01-24 | | Release date: | 2006-07-18 | | Last modified: | 2024-11-20 | | Method: | X-RAY DIFFRACTION (3.05 Å) | | Cite: | The Crystal Structure of Beryllofluoride Spo0F in Complex with the Phosphotransferase Spo0B Represents a Phosphotransfer Pretransition State.
J.Bacteriol., 188, 2006
|
|
3UIM
 
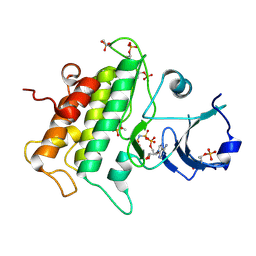 | |
3UNN
 
 | |
3UMZ
 
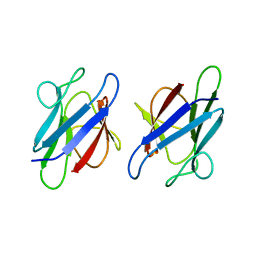 | | Crystal Structure of the human MDC1 FHA Domain | | Descriptor: | Mediator of DNA damage checkpoint protein 1 | | Authors: | Luo, S, Ye, K. | | Deposit date: | 2011-11-15 | | Release date: | 2012-01-25 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (1.65 Å) | | Cite: | Structural mechanism of the phosphorylation-dependent dimerization of the MDC1 forkhead-associated domain
Nucleic Acids Res., 40, 2012
|
|
2GMX
 
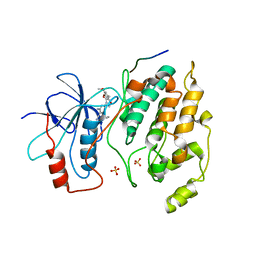 | | Selective Aminopyridine-Based C-Jun N-terminal Kinase inhibitors with cellular activity | | Descriptor: | C-jun-amino-terminal kinase-interacting protein 1, Mitogen-activated protein kinase 8, N-(4-AMINO-5-CYANO-6-ETHOXYPYRIDIN-2-YL)-2-(4-BROMO-2,5-DIMETHOXYPHENYL)ACETAMIDE, ... | | Authors: | Abad-Zapatero, C. | | Deposit date: | 2006-04-07 | | Release date: | 2006-06-06 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (3.5 Å) | | Cite: | Aminopyridine-Based c-Jun N-Terminal Kinase Inhibitors with Cellular Activity and Minimal Cross-Kinase Activity.
J.Med.Chem., 49, 2006
|
|
5GTR
 
 | |
9U9C
 
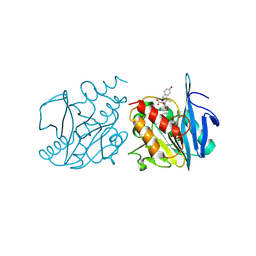 | | Crystal structure of NDM-1 in complex with hydrolyzed amoxicillin | | Descriptor: | (2~{R},4~{S})-2-[(1~{R})-1-[[(2~{R})-2-azanyl-2-(4-hydroxyphenyl)ethanoyl]amino]-2-oxidanyl-2-oxidanylidene-ethyl]-5,5-dimethyl-1,3-thiazolidine-4-carboxylic acid, Metallo-beta-lactamase type 2, ZINC ION | | Authors: | Shi, X, Zhang, Q, Liu, W. | | Deposit date: | 2025-03-27 | | Release date: | 2025-07-09 | | Method: | X-RAY DIFFRACTION (1.25 Å) | | Cite: | Crystal structure reveals the hydrophilic R1 group impairs NDM-1-ligand binding via water penetration at L3
J Struct Biol X, 2025
|
|
5WUD
 
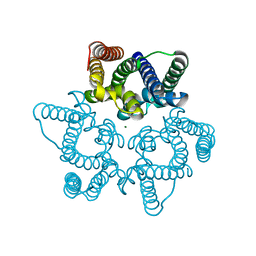 | | Structural basis for conductance through TRIC cation channels | | Descriptor: | MAGNESIUM ION, Uncharacterized protein | | Authors: | Su, M, Gao, F, Mao, Y, Li, D.L, Guo, Y.Z, Wang, X.H, Bruni, R, Kloss, B, Hendrickson, W.A, Chen, Y.H, New York Consortium on Membrane Protein Structure (NYCOMPS) | | Deposit date: | 2016-12-17 | | Release date: | 2017-06-21 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Structural basis for conductance through TRIC cation channels.
Nat Commun, 8, 2017
|
|
5IVX
 
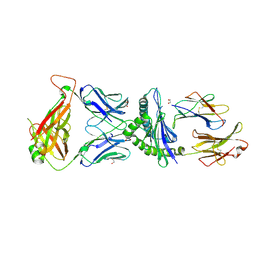 | | Crystal Structure of B4.2.3 T-Cell Receptor and H2-Dd P18-I10 Complex | | Descriptor: | 1,2-ETHANEDIOL, Beta-2-microglobulin, H-2 class I histocompatibility antigen, ... | | Authors: | Natarajan, K, Jiang, J, Margulies, D. | | Deposit date: | 2016-03-21 | | Release date: | 2017-03-29 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | An allosteric site in the T-cell receptor C beta domain plays a critical signalling role.
Nat Commun, 8, 2017
|
|
5IW1
 
 | |
5WUE
 
 | | Structural basis for conductance through TRIC cation channels | | Descriptor: | SULFATE ION, Uncharacterized protein | | Authors: | Su, M, Gao, F, Mao, Y, Li, D.L, Guo, Y.Z, Wang, X.H, Bruni, R, Kloss, B, Hendrickson, W.A, Chen, Y.H, New York Consortium on Membrane Protein Structure (NYCOMPS) | | Deposit date: | 2016-12-17 | | Release date: | 2017-06-21 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Structural basis for conductance through TRIC cation channels.
Nat Commun, 8, 2017
|
|
5WUF
 
 | | Structural basis for conductance through TRIC cation channels | | Descriptor: | CADMIUM ION, Putative membrane protein | | Authors: | Mao, Y, Gao, F, Su, M, Wang, X.H, Zeng, Y, Bruni, R, Kloss, B, Hendrickson, W.A, Chen, Y.H, New York Consortium on Membrane Protein Structure (NYCOMPS) | | Deposit date: | 2016-12-17 | | Release date: | 2017-08-09 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (2.401 Å) | | Cite: | Structural basis for conductance through TRIC cation channels.
Nat Commun, 8, 2017
|
|
5WUC
 
 | | Structural basis for conductance through TRIC cation channels | | Descriptor: | SODIUM ION, Uncharacterized protein | | Authors: | Su, M, Gao, F, Mao, Y, Li, D.L, Guo, Y.Z, Wang, X.H, Bruni, R, Kloss, B, Hendrickson, W.A, Chen, Y.H, New York Consortium on Membrane Protein Structure (NYCOMPS) | | Deposit date: | 2016-12-17 | | Release date: | 2017-07-12 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Structural basis for conductance through TRIC cation channels.
Nat Commun, 8, 2017
|
|
