7CZP
 
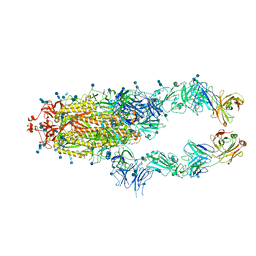 | | S protein of SARS-CoV-2 in complex bound with P2B-1A1 | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, IG c181_light_IGLV2-14_IGLJ3,IGL@ protein, ... | | Authors: | Yan, R.H, Zhang, Y.Y, Li, Y.N, Zhou, Q. | | Deposit date: | 2020-09-09 | | Release date: | 2021-03-10 | | Last modified: | 2024-10-16 | | Method: | ELECTRON MICROSCOPY (3 Å) | | Cite: | Structural basis for bivalent binding and inhibition of SARS-CoV-2 infection by human potent neutralizing antibodies.
Cell Res., 31, 2021
|
|
7CZQ
 
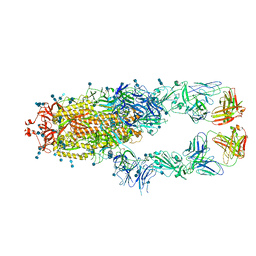 | | S protein of SARS-CoV-2 in complex bound with P2B-1A10 | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, IG c642_heavy_IGHV3-53_IGHD1-26_IGHJ6,Chain H of P2B-1A10,Immunoglobulin gamma-1 heavy chain, ... | | Authors: | Yan, R.H, Zhang, Y.Y, Li, Y.N, Zhou, Q. | | Deposit date: | 2020-09-09 | | Release date: | 2021-03-10 | | Last modified: | 2024-10-23 | | Method: | ELECTRON MICROSCOPY (2.8 Å) | | Cite: | Structural basis for bivalent binding and inhibition of SARS-CoV-2 infection by human potent neutralizing antibodies.
Cell Res., 31, 2021
|
|
7CZR
 
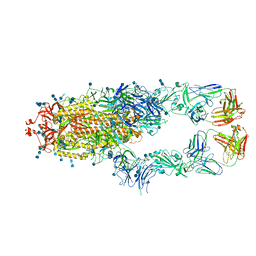 | | S protein of SARS-CoV-2 in complex bound with P5A-1B8_2B | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, IG c542_heavy_IGHV3-53_IGHD3-10_IGHJ6,IGH@ protein, ... | | Authors: | Yan, R.H, Zhang, Y.Y, Li, Y.N, Zhou, Q. | | Deposit date: | 2020-09-09 | | Release date: | 2021-03-10 | | Last modified: | 2024-10-30 | | Method: | ELECTRON MICROSCOPY (3.5 Å) | | Cite: | Structural basis for bivalent binding and inhibition of SARS-CoV-2 infection by human potent neutralizing antibodies.
Cell Res., 31, 2021
|
|
7CZT
 
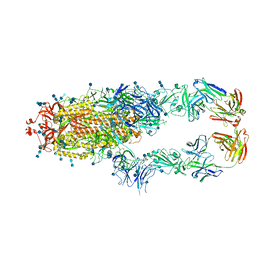 | | S protein of SARS-CoV-2 in complex bound with P5A-2G9 | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, IG c689_light_IGLV5-37_IGLJ3,IGL@ protein, ... | | Authors: | Yan, R.H, Zhang, Y.Y, Li, Y.N, Zhou, Q. | | Deposit date: | 2020-09-09 | | Release date: | 2021-03-10 | | Last modified: | 2024-11-06 | | Method: | ELECTRON MICROSCOPY (2.7 Å) | | Cite: | Structural basis for bivalent binding and inhibition of SARS-CoV-2 infection by human potent neutralizing antibodies.
Cell Res., 31, 2021
|
|
8ZYM
 
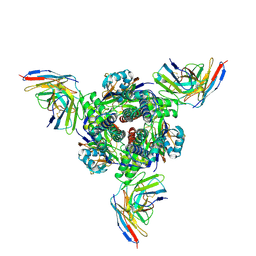 | | Complex structure of 60 Fab bound to DS2 prefusion F trimer | | Descriptor: | Fusion glycoprotein F0,Expression tag, antibody light chain, antidody heavy chain | | Authors: | Wang, X, Ge, J, Guo, F. | | Deposit date: | 2024-06-18 | | Release date: | 2025-04-30 | | Method: | ELECTRON MICROSCOPY (3.23 Å) | | Cite: | DS2 designer pre-fusion F vaccine induces strong and protective antibody response against RSV infection.
Npj Vaccines, 9, 2024
|
|
6JWE
 
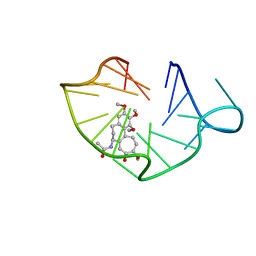 | | structure of RET G-quadruplex in complex with colchicine | | Descriptor: | DNA (5'-D(*GP*GP*GP*GP*CP*GP*GP*GP*GP*CP*GP*GP*GP*GP*CP*GP*GP*GP*GP*T)-3'), N-[(7S)-1,2,3,10-tetramethoxy-9-oxo-6,7-dihydro-5H-benzo[d]heptalen-7-yl]ethanamide | | Authors: | Wang, F, Wang, C, Liu, Y, Lan, W.X, Li, Y.M, Wang, R.X, Cao, C. | | Deposit date: | 2019-04-20 | | Release date: | 2020-02-26 | | Last modified: | 2024-05-15 | | Method: | SOLUTION NMR | | Cite: | Colchicine selective interaction with oncogene RET G-quadruplex revealed by NMR.
Chem.Commun.(Camb.), 56, 2020
|
|
6JWD
 
 | | structure of RET G-quadruplex in complex with berberine | | Descriptor: | BERBERINE, DNA (5'-D(*GP*GP*GP*GP*CP*GP*GP*GP*GP*CP*GP*GP*GP*GP*CP*GP*GP*GP*GP*T)-3') | | Authors: | Wang, F, Wang, C, Liu, Y, Lan, W.X, Li, Y.M, Wang, R.X, Cao, C. | | Deposit date: | 2019-04-19 | | Release date: | 2020-04-22 | | Last modified: | 2024-05-15 | | Method: | SOLUTION NMR | | Cite: | Colchicine selective interaction with oncogene RET G-quadruplex revealed by NMR.
Chem.Commun.(Camb.), 56, 2020
|
|
5Z0S
 
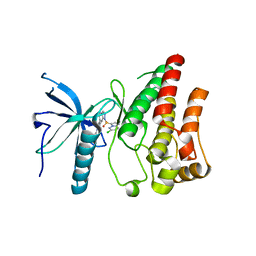 | | Crystal structure of FGFR1 kinase domain in complex with a novel inhibitor | | Descriptor: | 1-[(6-chloroimidazo[1,2-b]pyridazin-3-yl)sulfonyl]-6-(1-methyl-1H-pyrazol-4-yl)-1H-pyrazolo[4,3-b]pyridine, Fibroblast growth factor receptor 1 | | Authors: | Liu, Q, Xu, Y. | | Deposit date: | 2017-12-20 | | Release date: | 2018-12-26 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (2.45 Å) | | Cite: | Structure-Based Discovery of a Series of 5H-Pyrrolo[2,3-b]pyrazine FGFR Kinase Inhibitors
Molecules, 23, 2018
|
|
8ZTQ
 
 | | Crystal structure of Sufu from Mycoplasma Pneumonia | | Descriptor: | Nitrogen fixation protein NifU, ZINC ION | | Authors: | Wang, W.M, Ma, D.Y, Gong, W.J, Yao, H, Liu, Y.H, Wang, H.F. | | Deposit date: | 2024-06-07 | | Release date: | 2024-12-11 | | Last modified: | 2024-12-18 | | Method: | X-RAY DIFFRACTION (2.889 Å) | | Cite: | The reduced interaction between SufS and SufU in Mycoplasma penetrans results in diminished sulfotransferase activity.
Int.J.Biol.Macromol., 284, 2024
|
|
8ZTP
 
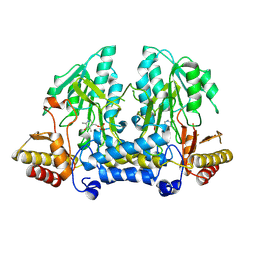 | | Crystal structure of cysteine desulfurase Sufs from Mycoplasma Pneumonia | | Descriptor: | PYRIDOXAL-5'-PHOSPHATE, cysteine desulfurase | | Authors: | Wang, W.M, Ma, D.Y, Gong, W.J, Yao, H, Liu, Y.H, Wang, H.F. | | Deposit date: | 2024-06-07 | | Release date: | 2024-12-11 | | Last modified: | 2024-12-18 | | Method: | X-RAY DIFFRACTION (3.2 Å) | | Cite: | The reduced interaction between SufS and SufU in Mycoplasma penetrans results in diminished sulfotransferase activity.
Int.J.Biol.Macromol., 284, 2024
|
|
9J3L
 
 | | ATP bound Arabidopsis ATP/ADP translocator AtNTT1 | | Descriptor: | ADENOSINE-5'-TRIPHOSPHATE, ADP,ATP carrier protein 1, chloroplastic, ... | | Authors: | Lin, H.J, Huang, J, Li, T.M, Li, W.J, Su, N.N, Zhang, J.R, Wu, X.D, Fan, M.R. | | Deposit date: | 2024-08-08 | | Release date: | 2025-03-19 | | Last modified: | 2025-05-21 | | Method: | ELECTRON MICROSCOPY (2.72 Å) | | Cite: | Structure and mechanism of the plastid/parasite ATP/ADP translocator.
Nature, 641, 2025
|
|
9J3M
 
 | | ADP/Pi bound Arabidopsis ATP/ADP translocator AtNTT1 | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, ADP,ATP carrier protein 1, chloroplastic, ... | | Authors: | Lin, H.J, Huang, J, Li, T.M, Li, W.J, Su, N.N, Zhang, J.R, Wu, X.D, Fan, M.R. | | Deposit date: | 2024-08-08 | | Release date: | 2025-03-19 | | Last modified: | 2025-05-21 | | Method: | ELECTRON MICROSCOPY (2.77 Å) | | Cite: | Structure and mechanism of the plastid/parasite ATP/ADP translocator.
Nature, 641, 2025
|
|
9J3O
 
 | | Chlamydia pneumoniae ATP/ADP translocator NTT1(Outward open state) | | Descriptor: | 1D10, ADP,ATP carrier protein 1 | | Authors: | Lin, H.J, Huang, J, Li, T.M, Li, W.J, Su, N.N, Zhang, J.R, Wu, X.D, Fan, M.R. | | Deposit date: | 2024-08-08 | | Release date: | 2025-03-19 | | Last modified: | 2025-05-21 | | Method: | ELECTRON MICROSCOPY (2.9 Å) | | Cite: | Structure and mechanism of the plastid/parasite ATP/ADP translocator.
Nature, 641, 2025
|
|
9J3J
 
 | | Arabidopsis ATP/ADP translocator AtNTT1 | | Descriptor: | ADP,ATP carrier protein 1, chloroplastic, nanobody: B-C8 | | Authors: | Lin, H.J, Huang, J, Li, T.M, Li, W.J, Su, N.N, Zhang, J.R, Wu, X.D, Fan, M.R. | | Deposit date: | 2024-08-08 | | Release date: | 2025-03-19 | | Last modified: | 2025-05-21 | | Method: | ELECTRON MICROSCOPY (2.83 Å) | | Cite: | Structure and mechanism of the plastid/parasite ATP/ADP translocator.
Nature, 641, 2025
|
|
9J3N
 
 | | ATP bound Chlamydia pneumoniae ATP/ADP translocator NTT1(Inward open state) | | Descriptor: | 1D10, ADENOSINE-5'-TRIPHOSPHATE, ADP,ATP carrier protein 1 | | Authors: | Lin, H.J, Huang, J, Li, T.M, Li, W.J, Su, N.N, Zhang, J.R, Wu, X.D, Fan, M.R. | | Deposit date: | 2024-08-08 | | Release date: | 2025-03-19 | | Last modified: | 2025-07-16 | | Method: | ELECTRON MICROSCOPY (2.72 Å) | | Cite: | Structure and mechanism of the plastid/parasite ATP/ADP translocator.
Nature, 641, 2025
|
|
9IQX
 
 | | Cryo-EM structure of the human TRPV4-RhoA in complex with AH001 | | Descriptor: | (1~{R})-1-(3-ethylphenyl)ethane-1,2-diol, GUANOSINE-5'-DIPHOSPHATE, MAGNESIUM ION, ... | | Authors: | Yuan, Z, Ruan, S.S, Li, S.L. | | Deposit date: | 2024-07-13 | | Release date: | 2025-05-28 | | Last modified: | 2025-07-02 | | Method: | ELECTRON MICROSCOPY (3.37 Å) | | Cite: | Inactivation of RhoA for Hypertension Treatment Through the TRPV4-RhoA-RhoGDI1 Axis.
Circulation, 2025
|
|
6PCH
 
 | | E. coli 50S ribosome bound to compound 21 | | Descriptor: | (3R,4R,5E,10E,12E,14S,26aR)-14-hydroxy-12-methyl-3-(propan-2-yl)-4-(prop-2-en-1-yl)-8,9,14,15,24,25,26,26a-octahydro-1H,3H,22H-21,18-(azeno)pyrrolo[2,1-c][1,8,4,19]dioxadiazacyclotetracosine-1,7,16,22(4H,17H)-tetrone, 23S ribosomal RNA, 50S ribosomal protein L13, ... | | Authors: | Pellegrino, J, Lee, D.J, Fraser, J.S, Seiple, I.B. | | Deposit date: | 2019-06-17 | | Release date: | 2020-06-17 | | Last modified: | 2024-12-25 | | Method: | ELECTRON MICROSCOPY (2.9 Å) | | Cite: | Synthetic group A streptogramin antibiotics that overcome Vat resistance.
Nature, 586, 2020
|
|
6JEP
 
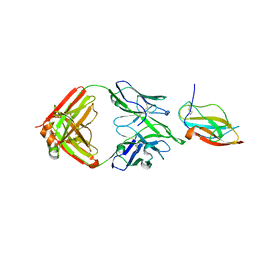 | | Structure of a neutralizing antibody bound to the Zika envelope protein domain III | | Descriptor: | Genome polyprotein, heavy chain of Fab ZK2B10, light chain of Fab ZK2B10 | | Authors: | Wang, L, Wang, R.K, Wang, L, Ben, H.J, Yu, L, Gao, F, Shi, X.L, Yin, C.B, Zhang, F.C, Xiang, Y, Zhang, L.Q. | | Deposit date: | 2019-02-07 | | Release date: | 2019-05-15 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (2.316 Å) | | Cite: | Structural Basis for Neutralization and Protection by a Zika Virus-Specific Human Antibody.
Cell Rep, 26, 2019
|
|
6PC7
 
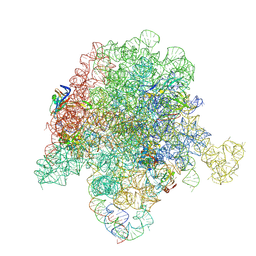 | | E. coli 50S ribosome bound to compound 46 | | Descriptor: | (2R)-2-[(3S,4R,5E,10E,12E,14S,16R,26aR)-16-fluoro-14-hydroxy-4,12-dimethyl-1,7,22-trioxo-4,7,8,9,14,15,16,17,24,25,26,26a-dodecahydro-1H,3H,22H-21,18-(azeno)pyrrolo[2,1-c][1,8,4,19]dioxadiazacyclotetracosin-3-yl]propyl isoquinolin-3-ylcarbamate, 23S ribosomal RNA, 50S ribosomal protein L13, ... | | Authors: | Pellegrino, J, Lee, D.J, Fraser, J.S, Seiple, I.B. | | Deposit date: | 2019-06-16 | | Release date: | 2020-06-17 | | Last modified: | 2024-12-25 | | Method: | ELECTRON MICROSCOPY (2.5 Å) | | Cite: | Synthetic group A streptogramin antibiotics that overcome Vat resistance.
Nature, 586, 2020
|
|
5Z1S
 
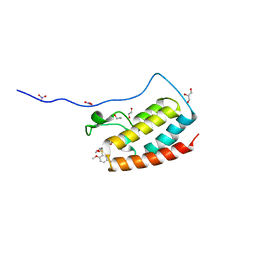 | | Crystal Structure Analysis of the BRD4(1) | | Descriptor: | 1,2-ETHANEDIOL, 5-bromo-2-methoxy-N-(6-methoxy-2,2-dimethyl-3-oxo-3,4-dihydro-2H-1,4-benzoxazin-7-yl)benzene-1-sulfonamide, Bromodomain-containing protein 4, ... | | Authors: | Xu, Y, Zhang, Y, Xiang, Q, Song, M, Wang, C. | | Deposit date: | 2017-12-28 | | Release date: | 2019-01-02 | | Last modified: | 2024-03-27 | | Method: | X-RAY DIFFRACTION (1.42 Å) | | Cite: | Y08060: A Selective BET Inhibitor for Treatment of Prostate Cancer.
Acs Med.Chem.Lett., 9, 2018
|
|
6PC8
 
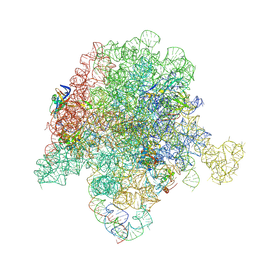 | | E. coli 50S ribosome bound to compound 40q | | Descriptor: | (2R)-2-[(3S,4R,5E,10E,12E,14S,26aR)-14-hydroxy-4,12-dimethyl-1,7,16,22-tetraoxo-4,7,8,9,14,15,16,17,24,25,26,26a-dodecahydro-1H,3H,22H-21,18-(azeno)pyrrolo[2,1-c][1,8,4,19]dioxadiazacyclotetracosin-3-yl]propyl isoquinolin-3-ylcarbamate, 23S ribosomal RNA, 50S ribosomal protein L13, ... | | Authors: | Pellegrino, J, Lee, D.J, Fraser, J.S, Seiple, I.B. | | Deposit date: | 2019-06-16 | | Release date: | 2020-06-17 | | Last modified: | 2024-12-25 | | Method: | ELECTRON MICROSCOPY (2.9 Å) | | Cite: | Synthetic group A streptogramin antibiotics that overcome Vat resistance.
Nature, 586, 2020
|
|
5Z1R
 
 | | Crystal Structure Analysis of the BRD4 | | Descriptor: | 1,2-ETHANEDIOL, 5-bromo-N-(2,2-dimethyl-3-oxo-3,4-dihydro-2H-1,4-benzoxazin-7-yl)-2-methoxybenzene-1-sulfonamide, Bromodomain-containing protein 4, ... | | Authors: | Xu, Y, Zhang, Y, Xiang, Q, Song, M, Wang, C. | | Deposit date: | 2017-12-28 | | Release date: | 2019-01-02 | | Last modified: | 2024-03-27 | | Method: | X-RAY DIFFRACTION (1.62 Å) | | Cite: | Y08060: A Selective BET Inhibitor for Treatment of Prostate Cancer.
Acs Med.Chem.Lett., 9, 2018
|
|
6PV9
 
 | |
8X82
 
 | |
8X83
 
 | |
