7DMY
 
 | | The crystal structure of Cpd7 in complex with BPTF bromodomain | | Descriptor: | Nucleosome-remodeling factor subunit BPTF, tert-butyl 3-methyl-2-[[(3R,5R)-1-methyl-5-phenyl-piperidin-3-yl]amino]-4-oxidanylidene-5,7-dihydropyrrolo[3,4-d]pyrimidine-6-carboxylate | | Authors: | Xiong, L, Guo, Y, Yang, S. | | Deposit date: | 2020-12-08 | | Release date: | 2021-10-20 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Discovery of selective BPTF bromodomain inhibitors by screening and structure-based optimization.
Biochem.Biophys.Res.Commun., 545, 2021
|
|
7DN4
 
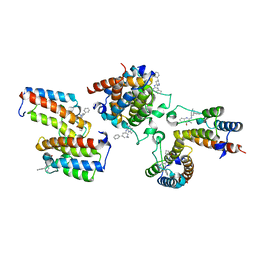 | | The crystal structure of Cpd8 in complex with BPTF bromodomain | | Descriptor: | 3-methyl-2-[[(3R,5R)-1-methyl-5-phenyl-piperidin-3-yl]amino]-6,7-dihydro-5H-cyclopenta[d]pyrimidin-4-one, Nucleosome-remodeling factor subunit BPTF | | Authors: | Xiong, L, Guo, Y, Yang, S. | | Deposit date: | 2020-12-08 | | Release date: | 2021-10-20 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2.841 Å) | | Cite: | Discovery of selective BPTF bromodomain inhibitors by screening and structure-based optimization.
Biochem.Biophys.Res.Commun., 545, 2021
|
|
7CD9
 
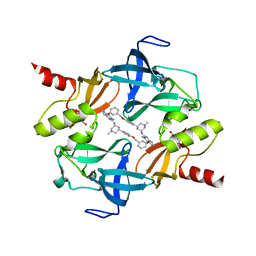 | | Crystal Structure of SETDB1 tudor domain in complexed with Compound 6 | | Descriptor: | 3-methyl-2-[[(3R,5R)-1-methyl-5-(4-phenylmethoxyphenyl)piperidin-3-yl]amino]-5H-pyrrolo[3,2-d]pyrimidin-4-one, CITRIC ACID, Histone-lysine N-methyltransferase SETDB1 | | Authors: | Xiong, L, Guo, Y, Mao, X, Huang, L, Wu, C, Yang, S. | | Deposit date: | 2020-06-19 | | Release date: | 2021-04-07 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Structure-Guided Discovery of a Potent and Selective Cell-Active Inhibitor of SETDB1 Tudor Domain.
Angew.Chem.Int.Ed.Engl., 60, 2021
|
|
4V7V
 
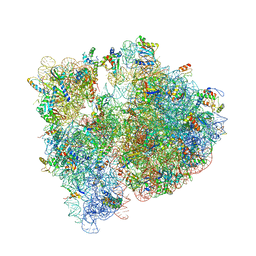 | | Crystal structure of the E. coli ribosome bound to clindamycin. | | Descriptor: | 16S rRNA, 23S rRNA, 30S ribosomal protein S10, ... | | Authors: | Dunkle, J.A, Xiong, L, Mankin, A.S, Cate, J.H.D. | | Deposit date: | 2010-08-16 | | Release date: | 2014-07-09 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (3.2891 Å) | | Cite: | Structures of the Escherichia coli ribosome with antibiotics bound near the peptidyl transferase center explain spectra of drug action.
Proc.Natl.Acad.Sci.USA, 107, 2010
|
|
4V7T
 
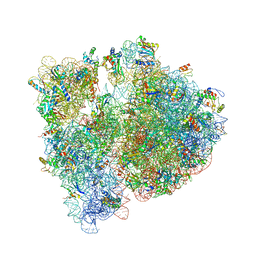 | | Crystal structure of the E. coli ribosome bound to chloramphenicol. | | Descriptor: | 16S rRNA, 23S rRNA, 30S ribosomal protein S10, ... | | Authors: | Dunkle, J.A, Xiong, L, Mankin, A.S, Cate, J.H.D. | | Deposit date: | 2010-08-14 | | Release date: | 2014-07-09 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (3.1942 Å) | | Cite: | Structures of the Escherichia coli ribosome with antibiotics bound near the peptidyl transferase center explain spectra of drug action.
Proc.Natl.Acad.Sci.USA, 107, 2010
|
|
4V7S
 
 | | Crystal structure of the E. coli ribosome bound to telithromycin. | | Descriptor: | 16S rRNA, 23S rRNA, 30S ribosomal protein S10, ... | | Authors: | Dunkle, J.A, Xiong, L, Mankin, A.S, Cate, J.H.D. | | Deposit date: | 2010-08-05 | | Release date: | 2014-07-09 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (3.2547 Å) | | Cite: | Structures of the Escherichia coli ribosome with antibiotics bound near the peptidyl transferase center explain spectra of drug action.
Proc.Natl.Acad.Sci.USA, 107, 2010
|
|
4V7U
 
 | | Crystal structure of the E. coli ribosome bound to erythromycin. | | Descriptor: | 16S rRNA, 23S rRNA, 30S ribosomal protein S10, ... | | Authors: | Dunkle, J.A, Xiong, L, Mankin, A.S, Cate, J.H.D. | | Deposit date: | 2010-08-15 | | Release date: | 2014-07-09 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (3.1 Å) | | Cite: | Structures of the Escherichia coli ribosome with antibiotics bound near the peptidyl transferase center explain spectra of drug action.
Proc.Natl.Acad.Sci.USA, 107, 2010
|
|
7C9N
 
 | | Crystal structure of SETDB1 tudor domain in complexed with Compound 1. | | Descriptor: | 3,5-dimethyl-2-[[(3R,5R)-1-methyl-5-phenyl-piperidin-3-yl]amino]pyrrolo[3,2-d]pyrimidin-4-one, Histone-lysine N-methyltransferase SETDB1 | | Authors: | Guo, Y, Xiong, L, Mao, X, Yang, S. | | Deposit date: | 2020-06-06 | | Release date: | 2021-04-07 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2.472 Å) | | Cite: | Structure-Guided Discovery of a Potent and Selective Cell-Active Inhibitor of SETDB1 Tudor Domain.
Angew.Chem.Int.Ed.Engl., 60, 2021
|
|
3WSQ
 
 | | Structure of HER2 with an Fab | | Descriptor: | Antibody Heavy Chain, Antibody Light Chain, Receptor tyrosine-protein kinase erbB-2 | | Authors: | Fu, W.Y, Wang, Y.X, Zhou, L.J. | | Deposit date: | 2014-03-20 | | Release date: | 2015-03-25 | | Last modified: | 2017-11-22 | | Method: | X-RAY DIFFRACTION (3.5 Å) | | Cite: | Insights into HER2 signaling from step-by-step optimization of anti-HER2 antibodies.
MAbs, 6, 2014
|
|
7OHA
 
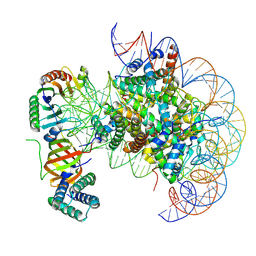 | | nucleosome with TBP and TFIIA bound at SHL +2 | | Descriptor: | DNA (122-MER), Histone H2A, Histone H2B 1.1, ... | | Authors: | Wang, H, Cramer, P. | | Deposit date: | 2021-05-09 | | Release date: | 2021-07-28 | | Last modified: | 2024-07-10 | | Method: | ELECTRON MICROSCOPY (2.9 Å) | | Cite: | Structures and implications of TBP-nucleosome complexes.
Proc.Natl.Acad.Sci.USA, 118, 2021
|
|
7OH9
 
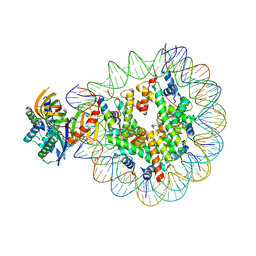 | | Nucleosome with TBP and TFIIA bound at SHL -6 | | Descriptor: | DNA (145-MER), Histone H2A, Histone H2B 1.1, ... | | Authors: | Wang, H, Cramer, P. | | Deposit date: | 2021-05-09 | | Release date: | 2021-07-28 | | Last modified: | 2024-07-10 | | Method: | ELECTRON MICROSCOPY (3 Å) | | Cite: | Structures and implications of TBP-nucleosome complexes.
Proc.Natl.Acad.Sci.USA, 118, 2021
|
|
7OHB
 
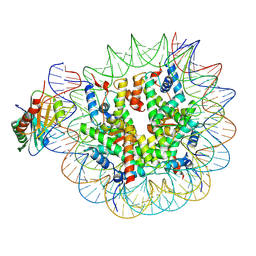 | | TBP-nucleosome complex | | Descriptor: | DNA (145-MER), Histone H2A, Histone H2B 1.1, ... | | Authors: | Wang, H, Cramer, P. | | Deposit date: | 2021-05-10 | | Release date: | 2021-07-28 | | Last modified: | 2024-07-10 | | Method: | ELECTRON MICROSCOPY (3.4 Å) | | Cite: | Structures and implications of TBP-nucleosome complexes.
Proc.Natl.Acad.Sci.USA, 118, 2021
|
|
7OHC
 
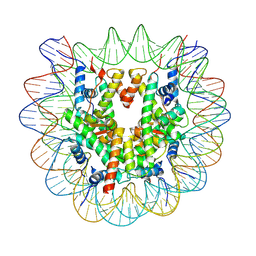 | |
3JQ4
 
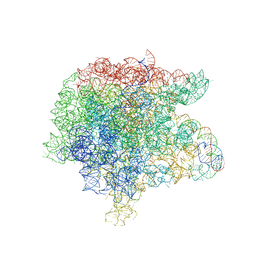 | | The structure of the complex of the large ribosomal subunit from D. Radiodurans with the antibiotic lankacidin | | Descriptor: | 23S ribosomal RNA, 5S ribosomal RNA, N-[(1S,2R,3E,5E,7S,9E,11E,13S,15R,19R)-7,13-dihydroxy-1,4,10,19-tetramethyl-17,18-dioxo-16-oxabicyclo[13.2.2]nonadeca-3,5,9,11-tetraen-2-yl]-2-oxopropanamide | | Authors: | Auerbach-Nevo, T, Mermershtain, I, Davidovich, C, Bashan, A, Rozenberg, H, Yonath, A. | | Deposit date: | 2009-09-06 | | Release date: | 2010-09-08 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (3.52 Å) | | Cite: | The structure of ribosome-lankacidin complex reveals ribosomal sites for synergistic antibiotics
Proc.Natl.Acad.Sci.USA, 107, 2010
|
|
4ZWB
 
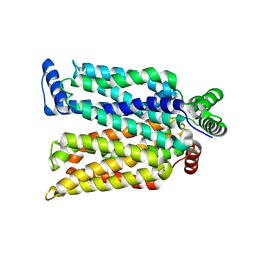 | | Crystal structure of maltose-bound human GLUT3 in the outward-occluded conformation at 2.4 angstrom | | Descriptor: | Solute carrier family 2, facilitated glucose transporter member 3, alpha-D-glucopyranose-(1-4)-alpha-D-glucopyranose | | Authors: | Deng, D, Sun, P.C, Yan, C.Y, Yan, N. | | Deposit date: | 2015-05-19 | | Release date: | 2015-07-22 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Molecular basis of ligand recognition and transport by glucose transporters
Nature, 526, 2015
|
|
4ZWC
 
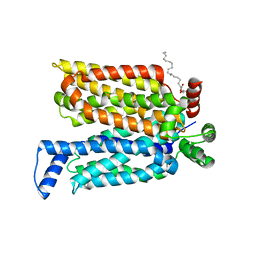 | | Crystal structure of maltose-bound human GLUT3 in the outward-open conformation at 2.6 angstrom | | Descriptor: | (2R)-2,3-dihydroxypropyl (9Z)-octadec-9-enoate, Solute carrier family 2, facilitated glucose transporter member 3, ... | | Authors: | Deng, D, Sun, P.C, Yan, C.Y, Yan, N. | | Deposit date: | 2015-05-19 | | Release date: | 2015-07-22 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Molecular basis of ligand recognition and transport by glucose transporters
Nature, 526, 2015
|
|
4ZW9
 
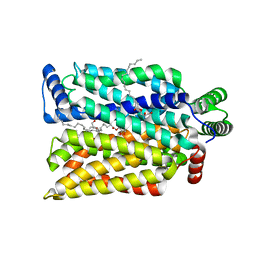 | | Crystal structure of human GLUT3 bound to D-glucose in the outward-occluded conformation at 1.5 angstrom | | Descriptor: | (2R)-2,3-dihydroxypropyl (9Z)-octadec-9-enoate, Solute carrier family 2, facilitated glucose transporter member 3, ... | | Authors: | Deng, D, Sun, P.C, Yan, C.Y, Yan, N. | | Deposit date: | 2015-05-19 | | Release date: | 2015-07-22 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (1.502 Å) | | Cite: | Molecular basis of ligand recognition and transport by glucose transporters
Nature, 526, 2015
|
|
3G43
 
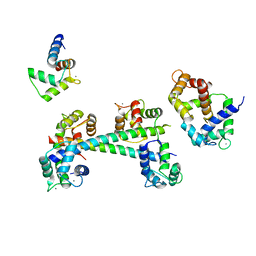 | |
7Y77
 
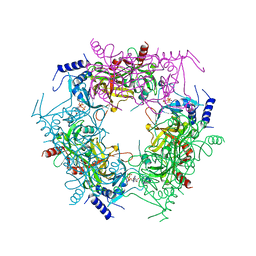 | | Crystal structure of rice NAL1 | | Descriptor: | ADENOSINE-5'-TRIPHOSPHATE, Protein NARROW LEAF 1 | | Authors: | Yan, J.J, Guan, Z.Y, Yin, P, Xiong, L.Z. | | Deposit date: | 2022-06-21 | | Release date: | 2023-07-12 | | Last modified: | 2024-05-29 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Serine protease NAL1 exerts pleiotropic functions through degradation of TOPLESS-related corepressor in rice.
Nat.Plants, 9, 2023
|
|
7CJT
 
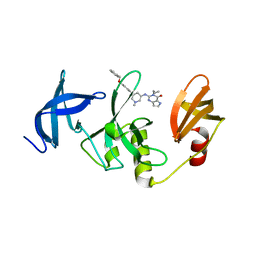 | | Crystal Structure of SETDB1 Tudor domain in complexed with (R,R)-59 | | Descriptor: | 2-[[(3~{R},5~{R})-1-methyl-5-(4-phenylmethoxyphenyl)piperidin-3-yl]amino]-3-prop-2-enyl-5~{H}-pyrrolo[3,2-d]pyrimidin-4-one, Histone-lysine N-methyltransferase SETDB1 | | Authors: | Guo, Y.P, Liang, X, Mao, X, Wu, C, Luyi, H, Yang, S. | | Deposit date: | 2020-07-13 | | Release date: | 2021-04-14 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2.474 Å) | | Cite: | Structure-Guided Discovery of a Potent and Selective Cell-Active Inhibitor of SETDB1 Tudor Domain.
Angew.Chem.Int.Ed.Engl., 60, 2021
|
|
7CAJ
 
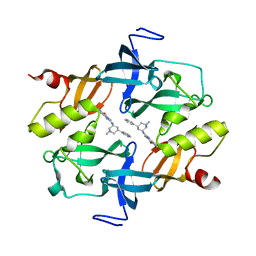 | | Crystal structure of SETDB1 Tudor domain in complexed with Compound 2. | | Descriptor: | 3-methyl-2-[[(3R,5R)-1-methyl-5-phenyl-piperidin-3-yl]amino]-5H-pyrrolo[3,2-d]pyrimidin-4-one, Histone-lysine N-methyltransferase SETDB1 | | Authors: | Guo, Y.P, Liang, X, Xin, M, Luyi, H, Chengyong, W, Yang, S.Y. | | Deposit date: | 2020-06-08 | | Release date: | 2021-04-07 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2.198 Å) | | Cite: | Structure-Guided Discovery of a Potent and Selective Cell-Active Inhibitor of SETDB1 Tudor Domain.
Angew.Chem.Int.Ed.Engl., 60, 2021
|
|
7X4E
 
 | | Structure of 10635-DndE | | Descriptor: | DNA sulfur modification protein DndE, GLYCEROL | | Authors: | Haiyan, G, Wei, H, Chen, S, Wang, L, Wu, G. | | Deposit date: | 2022-03-02 | | Release date: | 2022-04-20 | | Last modified: | 2024-05-29 | | Method: | X-RAY DIFFRACTION (2.32 Å) | | Cite: | Structural and Functional Analysis of DndE Involved in DNA Phosphorothioation in the Haloalkaliphilic Archaea Natronorubrum bangense JCM10635.
Mbio, 13, 2022
|
|
