6ES3
 
 | | Structure of CDX2-DNA(TCG) | | Descriptor: | DNA (5'-D(P*GP*GP*AP*GP*GP*TP*CP*GP*TP*AP*AP*AP*AP*CP*AP*CP*AP*A)-3'), DNA (5'-D(P*TP*TP*GP*TP*GP*TP*TP*TP*TP*AP*CP*GP*AP*CP*CP*TP*CP*C)-3'), Homeobox protein CDX-2 | | Authors: | Morgunova, E, Yin, Y, Jolma, A, Popov, A, Taipale, J. | | Deposit date: | 2017-10-19 | | Release date: | 2018-03-21 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (2.57 Å) | | Cite: | Two distinct DNA sequences recognized by transcription factors represent enthalpy and entropy optima.
Elife, 7, 2018
|
|
6ES2
 
 | | Structure of CDX2-DNA(CAA) | | Descriptor: | DNA (5'-D(P*GP*GP*AP*GP*GP*CP*AP*AP*TP*AP*AP*AP*AP*CP*AP*CP*AP*A)-3'), DNA (5'-D(P*TP*TP*GP*TP*GP*TP*TP*TP*TP*AP*TP*TP*GP*CP*CP*TP*CP*C)-3'), Homeobox protein CDX-2 | | Authors: | Morgunova, E, Yin, Y, Jolma, A, Popov, A, Taipale, J. | | Deposit date: | 2017-10-19 | | Release date: | 2018-03-21 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (2.95 Å) | | Cite: | Two distinct DNA sequences recognized by transcription factors represent enthalpy and entropy optima.
Elife, 7, 2018
|
|
2H3V
 
 | | Structure of the HIV-1 Matrix protein bound to di-C8-phosphatidylinositol-(4,5)-bisphosphate | | Descriptor: | Gag polyprotein, [(2R)-2-octanoyloxy-3-[oxidanyl-[(1R,2R,3S,4R,5R,6S)-2,3,6-tris(oxidanyl)-4,5-diphosphonooxy-cyclohexyl]oxy-phosphoryl]oxy-propyl] octanoate | | Authors: | Saad, J.S, Miller, J, Tai, J, Kim, A, Ghanam, R.H, Summers, M.F. | | Deposit date: | 2006-05-23 | | Release date: | 2006-07-25 | | Last modified: | 2024-05-29 | | Method: | SOLUTION NMR | | Cite: | Structural basis for targeting HIV-1 Gag proteins to the plasma membrane for virus assembly.
Proc.Natl.Acad.Sci.Usa, 103, 2006
|
|
2H3I
 
 | | Solution structure of the HIV-1 myristoylated Matrix protein | | Descriptor: | Gag polyprotein, MYRISTIC ACID | | Authors: | Saad, J.S, Miller, J, Tai, J, Kim, A, Ghanam, R.H, Summers, M.F. | | Deposit date: | 2006-05-22 | | Release date: | 2006-07-25 | | Last modified: | 2022-03-09 | | Method: | SOLUTION NMR | | Cite: | Structural basis for targeting HIV-1 Gag proteins to the plasma membrane for virus assembly.
Proc.Natl.Acad.Sci.Usa, 103, 2006
|
|
2H3Z
 
 | | Structure of the HIV-1 matrix protein bound to di-C4-phosphatidylinositol-(4,5)-bisphosphate | | Descriptor: | (2R)-3-{[(R)-HYDROXY{[(1R,2R,3S,4R,5R,6S)-2,3,6-TRIHYDROXY-4,5-BIS(PHOSPHONOOXY)CYCLOHEXYL]OXY}PHOSPHORYL]OXY}PROPANE-1 ,2-DIYL DIBUTANOATE, Gag polyprotein | | Authors: | Saad, J.S, Miller, J, Tai, J, Kim, A, Ghanam, R.H, Summers, M.F. | | Deposit date: | 2006-05-23 | | Release date: | 2006-07-25 | | Last modified: | 2024-05-01 | | Method: | SOLUTION NMR | | Cite: | Structural basis for targeting HIV-1 Gag proteins to the plasma membrane for virus assembly.
Proc.Natl.Acad.Sci.Usa, 103, 2006
|
|
2H3Q
 
 | | Solution structure of HIV-1 myrMA bound to di-C4-phosphatidylinositol-(4,5)-bisphosphate | | Descriptor: | (2R)-3-{[(R)-HYDROXY{[(1R,2R,3S,4R,5R,6S)-2,3,6-TRIHYDROXY-4,5-BIS(PHOSPHONOOXY)CYCLOHEXYL]OXY}PHOSPHORYL]OXY}PROPANE-1 ,2-DIYL DIBUTANOATE, Gag polyprotein, MYRISTIC ACID | | Authors: | Saad, J.S, Miller, J, Tai, J, Kim, A, Ghanam, R.H, Summers, M.F. | | Deposit date: | 2006-05-22 | | Release date: | 2006-07-25 | | Last modified: | 2024-10-30 | | Method: | SOLUTION NMR | | Cite: | Structural basis for targeting HIV-1 Gag proteins to the plasma membrane for virus assembly.
Proc.Natl.Acad.Sci.Usa, 103, 2006
|
|
2H3F
 
 | | Solution structure of the HIV-1 MA protein | | Descriptor: | Gag polyprotein | | Authors: | Saad, J.S, Miller, J, Tai, J, Kim, A, Ghanam, R.H, Summers, M.F. | | Deposit date: | 2006-05-22 | | Release date: | 2006-07-25 | | Last modified: | 2024-05-29 | | Method: | SOLUTION NMR | | Cite: | Structural basis for targeting HIV-1 Gag proteins to the plasma membrane for virus assembly.
Proc.Natl.Acad.Sci.Usa, 103, 2006
|
|
2JMG
 
 | | Solution structure of V7R mutant of HIV-1 myristoylated matrix protein | | Descriptor: | Gag polyprotein, MYRISTIC ACID | | Authors: | Saad, J.S, Loeliger, E, Luncsford, P, Liriano, M, Tai, J, Kim, A, Miller, J, Joshi, A, Freed, E.O, Summers, M.F. | | Deposit date: | 2006-11-11 | | Release date: | 2007-02-06 | | Last modified: | 2023-12-20 | | Method: | SOLUTION NMR | | Cite: | Point mutations in the HIV-1 matrix protein turn off the myristyl switch.
J.Mol.Biol., 366, 2007
|
|
2NV3
 
 | | Solution structure of L8A mutant of HIV-1 myristoylated matrix protein | | Descriptor: | Gag polyprotein, MYRISTIC ACID | | Authors: | Saad, J.S, Loeliger, E, Luncsford, P, Liriano, M, Tai, J, Kim, A, Miller, J, Joshi, A, Freed, E.O, Summers, M.F. | | Deposit date: | 2006-11-10 | | Release date: | 2007-02-06 | | Last modified: | 2024-10-30 | | Method: | SOLUTION NMR | | Cite: | Point mutations in the HIV-1 matrix protein turn off the myristyl switch.
J.Mol.Biol., 366, 2007
|
|
6M7L
 
 | | Complex of OxyA with the X-domain from GPA biosynthesis | | Descriptor: | PROTOPORPHYRIN IX CONTAINING FE, Putative cytochrome P450 hydroxylase, Putative non-ribosomal peptide synthetase | | Authors: | Greule, A, Izore, T, Tailhades, J, Peschke, M, Schoppet, M, Ahmed, I, Kulik, A, Adamek, M, Ziemert, N, De Voss, J, Stegmann, E, Cryle, M.J. | | Deposit date: | 2018-08-20 | | Release date: | 2019-05-22 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (2.648297 Å) | | Cite: | Kistamicin biosynthesis reveals the biosynthetic requirements for production of highly crosslinked glycopeptide antibiotics.
Nat Commun, 10, 2019
|
|
5LUX
 
 | | Homeobox transcription factor CDX1 bound to methylated DNA | | Descriptor: | DNA (5'-D(P*GP*AP*GP*GP*TP*(5CM)P*GP*TP*AP*AP*AP*AP*CP*AP*CP*AP*A)-3'), DNA (5'-D(P*GP*GP*AP*GP*GP*TP*(5CM)P*GP*TP*AP*AP*AP*AP*CP*AP*CP*AP*A)-3'), DNA (5'-D(P*TP*TP*GP*TP*GP*TP*TP*TP*TP*AP*(5CM)P*GP*AP*CP*CP*TP*C)-3'), ... | | Authors: | Morgunova, E, Popov, A, Taipale, J. | | Deposit date: | 2016-09-12 | | Release date: | 2017-05-17 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (3.23 Å) | | Cite: | Impact of cytosine methylation on DNA binding specificities of human transcription factors.
Science, 356, 2017
|
|
5LTY
 
 | | Homeobox transcription factor CDX2 bound to methylated DNA | | Descriptor: | DNA (5'-D(P*GP*GP*AP*GP*GP*TP*(5CM)P*GP*TP*AP*AP*AP*AP*CP*AP*CP*AP*A)-3'), DNA (5'-D(P*TP*TP*GP*TP*GP*TP*TP*TP*TP*AP*(5CM)P*GP*AP*CP*CP*TP*CP*C)-3'), Homeobox protein CDX-2 | | Authors: | Morgunova, E, Popov, A, Taipale, J. | | Deposit date: | 2016-09-07 | | Release date: | 2017-05-31 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (2.66 Å) | | Cite: | Impact of cytosine methylation on DNA binding specificities of human transcription factors.
Science, 356, 2017
|
|
4XRM
 
 | | homodimer of TALE type homeobox transcription factor MEIS1 complexes with specific DNA | | Descriptor: | 2-AMINO-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, DNA (5'-D(P*AP*GP*CP*TP*GP*AP*CP*AP*GP*CP*TP*GP*TP*CP*AP*AP*G)-3'), DNA (5'-D(P*TP*CP*TP*TP*GP*AP*CP*AP*GP*CP*TP*GP*TP*CP*AP*GP*C)-3'), ... | | Authors: | Morgunova, E, Jorma, A, Yin, Y, Nitta, K.R, Dave, K, Enge, M, Kivioja, T, Popov, A, Taipale, J. | | Deposit date: | 2015-01-21 | | Release date: | 2015-11-04 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | DNA-dependent formation of transcription factor pairs alters their binding specificity.
Nature, 527, 2015
|
|
4XRS
 
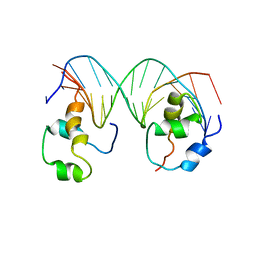 | | Heterodimeric complex of transcription factors MEIS1 and DLX3 on specific DNA | | Descriptor: | DNA (5'-D(P*AP*CP*AP*AP*TP*TP*AP*TP*CP*CP*TP*GP*TP*CP*AP*AP*C)-3'), DNA (5'-D(P*CP*AP*AP*TP*TP*AP*TP*CP*CP*TP*GP*TP*CP*AP*A)-3'), DNA (5'-D(P*GP*TP*TP*GP*AP*CP*AP*GP*GP*AP*TP*AP*AP*TP*TP*GP*TP*T)-3'), ... | | Authors: | Jorma, A, Yin, Y, Nitta, K.R, Dave, K, Enge, M, Kivioja, T, Popov, A, Morgunova, E, Taipale, J. | | Deposit date: | 2015-01-21 | | Release date: | 2015-11-04 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (3.5 Å) | | Cite: | DNA-dependent formation of transcription factor pairs alters their binding specificity.
Nature, 527, 2015
|
|
8OSB
 
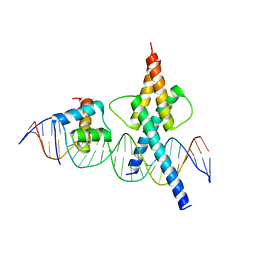 | | TWIST1-TCF4-ALX4 complex on specific DNA | | Descriptor: | DNA (25-MER), Homeobox protein aristaless-like 4, Transcription factor 4, ... | | Authors: | Morgunova, E, Kim, S, Popov, A, Wysocka, J, Taipale, J. | | Deposit date: | 2023-04-18 | | Release date: | 2024-01-31 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (2.9 Å) | | Cite: | DNA-guided transcription factor cooperativity shapes face and limb mesenchyme.
Cell, 187, 2024
|
|
5HOD
 
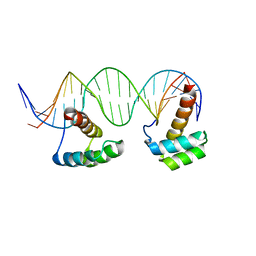 | | Structure of LHX4 transcription factor complexed with DNA | | Descriptor: | DNA (5'-D(P*AP*CP*CP*TP*AP*AP*TP*TP*AP*GP*GP*CP*GP*TP*AP*AP*TP*TP*AP*G)-3'), DNA (5'-D(P*CP*TP*AP*AP*TP*TP*AP*CP*GP*CP*CP*TP*AP*AP*TP*TP*AP*GP*GP*T)-3'), LIM/homeobox protein Lhx4 | | Authors: | Morgunova, E, Yin, Y, Popov, A, Taipale, J. | | Deposit date: | 2016-01-19 | | Release date: | 2017-02-01 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (2.682 Å) | | Cite: | Impact of cytosine methylation on DNA binding specificities of human transcription factors.
Science, 356, 2017
|
|
5EGO
 
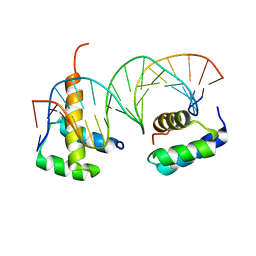 | | HOXB13-MEIS1 heterodimer bound to methylated DNA | | Descriptor: | DNA (5'-D(*CP*CP*TP*(5CM)P*GP*TP*AP*AP*AP*AP*CP*TP*GP*TP*CP*AP*AP*C)-3'), DNA (5'-D(P*GP*TP*TP*GP*AP*CP*AP*GP*TP*TP*TP*TP*AP*(5CM)P*GP*AP*GP*G)-3'), Homeobox protein Hox-B13, ... | | Authors: | Morgunova, E, Yin, Y, Jolma, A, Popov, A, Taipale, J. | | Deposit date: | 2015-10-27 | | Release date: | 2016-11-09 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (2.54 Å) | | Cite: | Impact of cytosine methylation on DNA binding specificities of human transcription factors.
Science, 356, 2017
|
|
5EF6
 
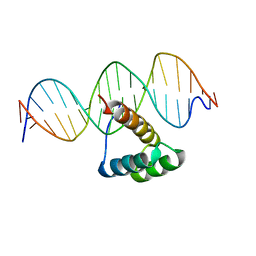 | | Structure of HOXB13 complex with methylated DNA | | Descriptor: | DNA (5'-D(P*GP*GP*AP*CP*CP*TP*(5CM)P*GP*TP*AP*AP*AP*AP*CP*AP*CP*AP*A)-3'), DNA (5'-D(P*TP*TP*GP*TP*GP*TP*TP*TP*TP*AP*(5CM)P*GP*AP*GP*GP*TP*CP*C)-3'), Homeobox protein Hox-B13 | | Authors: | Morgunova, E, Yin, Y, Jolma, A, Popov, A, Taipale, J. | | Deposit date: | 2015-10-23 | | Release date: | 2017-02-08 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (3 Å) | | Cite: | Impact of cytosine methylation on DNA binding specificities of human transcription factors.
Science, 356, 2017
|
|
8PNC
 
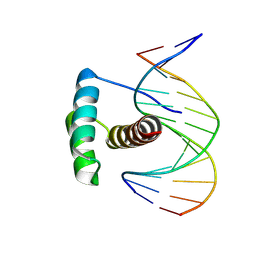 | |
5NNX
 
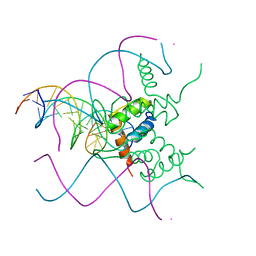 | | TEAD1 bound to DNA | | Descriptor: | DNA, Transcriptional enhancer factor TEF-1 | | Authors: | Morgunova, E, Jolma, A, Yin, Y, Popov, A, Taipale, J. | | Deposit date: | 2017-04-10 | | Release date: | 2017-04-26 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (3.29 Å) | | Cite: | TEAD1 bound to DNA
To Be Published
|
|
7PSX
 
 | | Structure of HOXB13 bound to hydroxymethylated DNA | | Descriptor: | DNA (5'-D(P*GP*GP*AP*CP*CP*TP*5HCP*AP*TP*AP*AP*AP*AP*CP*AP*CP*AP*A)-3'), DNA (5'-D(P*TP*TP*GP*TP*GP*TP*TP*TP*TP*AP*CP*GP*AP*GP*GP*TP*CP*C)-3'), Homeobox protein Hox-B13, ... | | Authors: | Morgunova, E, Popov, A, Yin, Y, Taipale, J. | | Deposit date: | 2021-09-24 | | Release date: | 2022-10-05 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Structure of HOXB13 bound to hydroxymethylated DNA
To Be Published
|
|
7Q3O
 
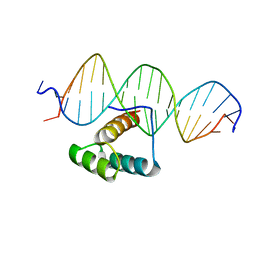 | |
5NO6
 
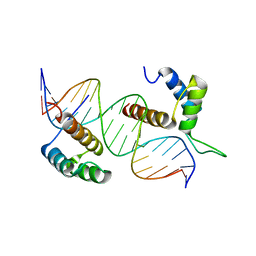 | | TEAD4-HOXB13 complex bound to DNA | | Descriptor: | DNA, Homeobox protein Hox-B13, Transcriptional enhancer factor TEF-3 | | Authors: | Morgunova, E, Jolma, A, Yin, Y, Popov, A, Taipale, J. | | Deposit date: | 2017-04-11 | | Release date: | 2018-05-16 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (2.88 Å) | | Cite: | TEAD4-HOXB13 complex bound to DNA
To Be Published
|
|
8PMF
 
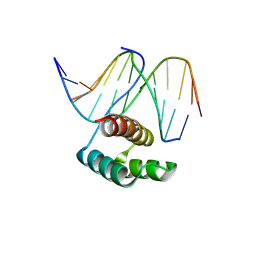 | |
8PMN
 
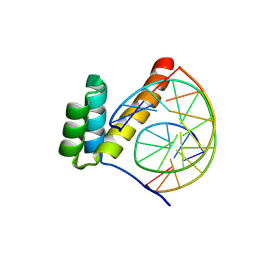 | |
