5OXE
 
 | |
2X6V
 
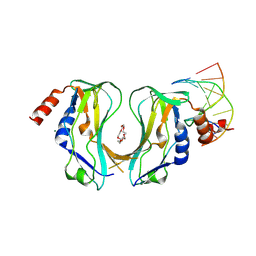 | | Crystal structure of human TBX5 in the DNA-bound and DNA-free form | | Descriptor: | 2-{2-[2-(2-{2-[2-(2-ETHOXY-ETHOXY)-ETHOXY]-ETHOXY}-ETHOXY)-ETHOXY]-ETHOXY}-ETHANOL, 5'-D(*TP*AP*AP*GP*GP*TP*GP*TP*GP*AP*GP)-3', 5'-D(*TP*CP*TP*CP*AP*CP*AP*CP*CP*TP*TP)-3', ... | | Authors: | Ptchelkine, D, Stirnimann, C.U, Grimm, C, Mueller, C.W. | | Deposit date: | 2010-02-22 | | Release date: | 2010-04-28 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Structural Basis of Tbx5-DNA Recognition: The T-Box Domain in its DNA-Bound and -Unbound Form.
J.Mol.Biol., 400, 2010
|
|
6QJS
 
 | | Crystal structure of a DNA dodecamer containing a tetramethylpiperidinoxyl (nitroxide) spin label | | Descriptor: | COBALT HEXAMMINE(III), DNA (5'-D(P*GP*CP*AP*AP*AP*TP*TP*(S6M)P*GP*CP*G)-3') | | Authors: | Hardwick, J.S, Haugland, M.M, Ptchelkine, D, Brown, T, Anderson, E.E. | | Deposit date: | 2019-01-24 | | Release date: | 2020-02-12 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | 2'-Alkynyl spin-labelling is a minimally perturbing tool for DNA structural analysis.
Nucleic Acids Res., 48, 2020
|
|
6QJR
 
 | | Crystal structure of a DNA dodecamer containing a tetramethylpyrrolinoxyl (nitroxide) spin label | | Descriptor: | DNA (5'-D(*CP*GP*CP*AP*AP*AP*AP*AP*AP*GP*CP*G)-3'), DNA (5'-D(*CP*GP*CP*TP*TP*TP*TP*TP*(5NO)P*GP*CP*G)-3') | | Authors: | Hardwick, J.S, Haugland, M.M, Ptchelkine, D, Brown, T, Anderson, E.E. | | Deposit date: | 2019-01-24 | | Release date: | 2020-02-12 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (2.9 Å) | | Cite: | 2'-Alkynyl spin-labelling is a minimally perturbing tool for DNA structural analysis.
Nucleic Acids Res., 48, 2020
|
|
2VY2
 
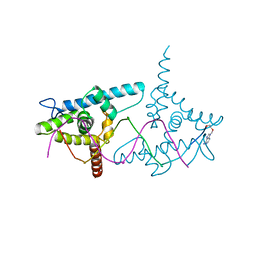 | | Structure of LEAFY transcription factor from Arabidopsis thaliana in complex with DNA from AG-I promoter | | Descriptor: | 5'-D(*AP*TP*TP*TP*AP*AP*TP*CP*CP*AP *AP*TP*GP*GP*TP*TP*AP*CP*AP*A)-3', PROTEIN LEAFY | | Authors: | Hames, C, Ptchelkine, D, Grimm, C, Thevenon, E, Moyroud, E, Gerard, F, Martiel, J.L, Benlloch, R, Parcy, F, Muller, C.W. | | Deposit date: | 2008-07-16 | | Release date: | 2008-09-23 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Structural Basis for Leafy Floral Switch Function and Similarity with Helix-Turn-Helix Proteins.
Embo J., 27, 2008
|
|
2VY1
 
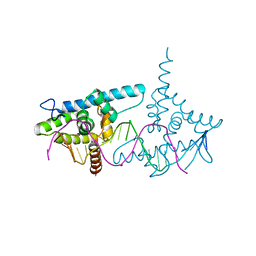 | | Structure of LEAFY transcription factor from Arabidopsis thaliana in complex with DNA from AP1 promoter | | Descriptor: | 5'-D(*TP*TP*AP*CP*GP*GP*AP*CP*CP*AP *CP*TP*GP*GP*TP*CP*CP*TP*TP*CP)-3', PROTEIN LEAFY | | Authors: | Hames, C, Ptchelkine, D, Grimm, C, Thevenon, E, Moyroud, E, Gerard, F, Martiel, J.L, Benlloch, R, Parcy, F, Muller, C.W. | | Deposit date: | 2008-07-16 | | Release date: | 2008-09-23 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (2.104 Å) | | Cite: | Structural Basis for Leafy Floral Switch Function and Similarity with Helix-Turn-Helix Proteins.
Embo J., 27, 2008
|
|
4ELC
 
 | | Crystal structure of the catalytic domain of botulinum neurotoxin BoNT/A C134 mutant with MTSEA modified Cys-165 | | Descriptor: | (2S)-2-hydroxybutanedioic acid, (4S)-2-METHYL-2,4-PENTANEDIOL, Botulinum neurotoxin A light chain, ... | | Authors: | Stura, E.A, Vera, L, Ptchelkine, D, Bakirci, H, Garcia, S, Dive, V. | | Deposit date: | 2012-04-10 | | Release date: | 2012-08-15 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Structural Framework for Covalent Inhibition of Clostridium botulinum Neurotoxin A by Targeting Cys165.
J.Biol.Chem., 287, 2012
|
|
4KUF
 
 | | Crystal structure of the catalytic domain of botulinum neurotoxin BoNT/A C134 mutant with MTSEA modified Cys-165 causing stretch disorder | | Descriptor: | (2S)-2-hydroxybutanedioic acid, 1,2-ETHANEDIOL, Botulinum neurotoxin A light chain, ... | | Authors: | Stura, E.A, Vera, L, Ptchelkine, D, Bakirci, H, Garcia, S, Dive, V. | | Deposit date: | 2013-05-22 | | Release date: | 2014-07-09 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (1.697 Å) | | Cite: | Covalent modification of the active site cysteine stresses Clostridium botulinum neurotoxin A
To be Published
|
|
4EL4
 
 | | Crystal structure of the catalytic domain of botulinum neurotoxin BoNT/A C134S/C165S double mutant | | Descriptor: | 1,2-ETHANEDIOL, Botulinum neurotoxin A light chain, GLYCEROL, ... | | Authors: | Stura, E.A, Vera, L, Ptchelkine, D, Dive, V. | | Deposit date: | 2012-04-10 | | Release date: | 2012-08-15 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (1.2 Å) | | Cite: | Structural Framework for Covalent Inhibition of Clostridium botulinum Neurotoxin A by Targeting Cys165.
J.Biol.Chem., 287, 2012
|
|
8ORH
 
 | | Knockout of GMC-oxidoreductase genes reveals that functional redundancy preserves mimivirus essential functions | | Descriptor: | FLAVIN-ADENINE DINUCLEOTIDE, Putative GMC-type oxidoreductase | | Authors: | Alempic, J.M, Bisio, H, Villalta, A, Santini, S, Lartigue, A, Schmitt, A, Bugnot, C, Notaro, A, Belmudes, L, Adrait, A, Poirot, O, Ptchelkine, D, De Castro, C, Coute, Y, Abergel, C. | | Deposit date: | 2023-04-14 | | Release date: | 2024-04-17 | | Last modified: | 2024-05-08 | | Method: | ELECTRON MICROSCOPY (4.2 Å) | | Cite: | Functional redundancy revealed by the deletion of the mimivirus GMC-oxidoreductase genes.
Microlife, 5, 2024
|
|
8ORS
 
 | | Knockout of GMC-oxidoreductase genes reveals that functional redundancy preserves mimivirus essential functions | | Descriptor: | FLAVIN-ADENINE DINUCLEOTIDE, Putative GMC-type oxidoreductase | | Authors: | Alempic, J.M, Bisio, H, Villalta, A, Santini, S, Lartigue, A, Schmitt, A, Bugnot, C, Notaro, A, Belmudes, L, Adrait, A, Poirot, O, Ptchelkine, D, De Castro, C, Coute, Y, Abergel, C. | | Deposit date: | 2023-04-17 | | Release date: | 2024-04-17 | | Last modified: | 2024-05-08 | | Method: | ELECTRON MICROSCOPY (4.3 Å) | | Cite: | Functional redundancy revealed by the deletion of the mimivirus GMC-oxidoreductase genes.
Microlife, 5, 2024
|
|
8PHZ
 
 | | Helical reconstruction of CHIKV nsP3 helical scaffolds | | Descriptor: | Non-structural protein 3, ZINC ION | | Authors: | Reguera, J, Hons, M, Zimberger, C, Ptchelkine, D, Jones, R, Desfosses, A. | | Deposit date: | 2023-06-20 | | Release date: | 2024-08-14 | | Method: | ELECTRON MICROSCOPY (2.35 Å) | | Cite: | The alphavirus nsP3 protein forms helical tubular scaffolds important for viral replication and particle assembly
To be published
|
|
8PK7
 
 | | Helical reconstruction of CHIKV nsP3 helical scaffolds | | Descriptor: | Non-structural protein 3, ZINC ION | | Authors: | Reguera, J, Hons, M, Zimberger, C, Ptchelkine, D, Jones, R, Desfosses, A. | | Deposit date: | 2023-06-25 | | Release date: | 2024-08-14 | | Method: | ELECTRON MICROSCOPY (2.52 Å) | | Cite: | The alphavirus nsP3 protein forms helical tubular scaffolds important for viral replication and particle assembly
To Be Published
|
|
5MVT
 
 | | Crystal structure of an A-DNA dodecamer featuring an alternating pyrimidine-purine sequence | | Descriptor: | COBALT (III) ION, DNA | | Authors: | Hardwick, J.S, Ptchelkine, D, Phillips, S.E.V, Brown, T. | | Deposit date: | 2017-01-17 | | Release date: | 2017-05-10 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (1.896 Å) | | Cite: | 5-Formylcytosine does not change the global structure of DNA.
Nat. Struct. Mol. Biol., 24, 2017
|
|
5MVQ
 
 | | Crystal structure of an unmodified, self-complementary dodecamer. | | Descriptor: | DNA, MAGNESIUM ION | | Authors: | Hardwick, J.S, Ptchelkine, D, Phillips, S.E.V, Brown, T. | | Deposit date: | 2017-01-17 | | Release date: | 2017-05-10 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (1.604 Å) | | Cite: | 5-Formylcytosine does not change the global structure of DNA.
Nat. Struct. Mol. Biol., 24, 2017
|
|
5MVU
 
 | |
5MVP
 
 | | Crystal structure of an A-DNA dodecamer containing the GGGCCC motif | | Descriptor: | DNA, POTASSIUM ION | | Authors: | Hardwick, J.S, Ptchelkine, D, Phillips, S.E.V, Brown, T. | | Deposit date: | 2017-01-17 | | Release date: | 2017-05-10 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (1.606 Å) | | Cite: | 5-Formylcytosine does not change the global structure of DNA.
Nat. Struct. Mol. Biol., 24, 2017
|
|
5MVK
 
 | |
5MVL
 
 | | Crystal structure of an A-DNA dodecamer containing 5-bromouracil | | Descriptor: | Brominated DNA dodecamer, MAGNESIUM ION | | Authors: | Hardwick, J.S, Ptchelkine, D, Phillips, S.E.V, Brown, T. | | Deposit date: | 2017-01-16 | | Release date: | 2017-05-10 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (1.405 Å) | | Cite: | 5-Formylcytosine does not change the global structure of DNA.
Nat. Struct. Mol. Biol., 24, 2017
|
|
5N88
 
 | | Crystal structure of antibody bound to viral protein | | Descriptor: | PC4 and SFRS1-interacting protein, VH59 antibody | | Authors: | Bao, L, Hannon, C, Cruz-Migoni, A, Ptchelkine, D, Sun, M.-y, Derveni, M, Bunjobpol, W, Chambers, J.S, Simmons, A, Phillips, S.E.V, Rabbitts, T.H. | | Deposit date: | 2017-02-23 | | Release date: | 2017-12-20 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Intracellular immunization against HIV infection with an intracellular antibody that mimics HIV integrase binding to the cellular LEDGF protein.
Sci Rep, 7, 2017
|
|
2X6U
 
 | | Crystal structure of human TBX5 in the DNA-free form | | Descriptor: | 2-{2-[2-(2-{2-[2-(2-ETHOXY-ETHOXY)-ETHOXY]-ETHOXY}-ETHOXY)-ETHOXY]-ETHOXY}-ETHANOL, MAGNESIUM ION, T-BOX TRANSCRIPTION FACTOR TBX5 | | Authors: | Stirnimann, C.U, Mueller, C.W. | | Deposit date: | 2010-02-22 | | Release date: | 2010-04-28 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Structural Basis of Tbx5-DNA Recognition: The T-Box Domain in its DNA-Bound and -Unbound Form.
J.Mol.Biol., 400, 2010
|
|
4EJ5
 
 | | Crystal structure of the catalytic domain of botulinum neurotoxin BoNT/A wild-type | | Descriptor: | 1,2-ETHANEDIOL, Botulinum neurotoxin A light chain, CARBONATE ION, ... | | Authors: | Stura, E.A, Vera, L, Dive, V. | | Deposit date: | 2012-04-06 | | Release date: | 2012-08-15 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (1.87 Å) | | Cite: | Structural Framework for Covalent Inhibition of Clostridium botulinum Neurotoxin A by Targeting Cys165.
J.Biol.Chem., 287, 2012
|
|
4KTX
 
 | | Crystal structure of the catalytic domain of botulinum neurotoxin BoNT/A C134S mutant with covalent inhibitor that modifies Cys-165 causing disorder in 167-174 stretch | | Descriptor: | Botulinum neurotoxin A light chain, GLYCEROL, Peptide inhibitor MPT-DPP-ARG-G-LEU-NH2, ... | | Authors: | Stura, E.A, Vera, L, Guitot, K, Dive, V. | | Deposit date: | 2013-05-21 | | Release date: | 2014-10-15 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (2.59 Å) | | Cite: | Covalent modification of the active site cysteine stresses Clostridium botulinum neurotoxin A
To be Published
|
|
4KS6
 
 | | Crystal structure of the catalytic domain of botulinum neurotoxin BoNT/A C134S mutant with covalent inhibitor that modifies Cys-165 causing disorder in 166-174 stretch | | Descriptor: | 1,2-ETHANEDIOL, Botulinum neurotoxin A light chain, DI(HYDROXYETHYL)ETHER, ... | | Authors: | Stura, E.A, Vera, L, Guitot, K, Dive, V. | | Deposit date: | 2013-05-17 | | Release date: | 2014-06-25 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (1.93 Å) | | Cite: | Covalent modification of the active site cysteine stresses Clostridium botulinum neurotoxin A
To be Published
|
|
