1QJP
 
 | |
1JQH
 
 | | IGF-1 receptor kinase domain | | Descriptor: | IGF-1 receptor kinase, MAGNESIUM ION, PHOSPHOAMINOPHOSPHONIC ACID-ADENYLATE ESTER, ... | | Authors: | Pautsch, A, Zoephel, A, Ahorn, H, Spevak, W, Hauptmann, R, Nar, H. | | Deposit date: | 2001-08-07 | | Release date: | 2002-04-19 | | Last modified: | 2024-12-25 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Crystal structure of bisphosphorylated IGF-1 receptor kinase: insight into domain movements upon kinase activation.
Structure, 9, 2001
|
|
2QLL
 
 | | Human liver glycogen phosphorylase- GL complex | | Descriptor: | Glycogen phosphorylase, liver form, PYRIDOXAL-5'-PHOSPHATE, ... | | Authors: | Pautsch, A, Streicher, R, Wissdorf, O, Stadler, N. | | Deposit date: | 2007-07-13 | | Release date: | 2008-02-19 | | Last modified: | 2025-03-26 | | Method: | X-RAY DIFFRACTION (2.56 Å) | | Cite: | Molecular recognition of the protein phosphatase 1 glycogen targeting subunit by glycogen phosphorylase.
J.Biol.Chem., 283, 2008
|
|
4BBA
 
 | | Crystal structure of glucokinase regulatory protein complexed to phosphate | | Descriptor: | GLUCOKINASE REGULATORY PROTEIN, PHOSPHATE ION | | Authors: | Pautsch, A, Stadler, N, Loehle, A, Lenter, M, Rist, W, Berg, A, Glocker, L, Nar, H, Reinhart, D, Heckel, A, Schnapp, G, Kauschke, S.G. | | Deposit date: | 2012-09-21 | | Release date: | 2013-05-15 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (1.92 Å) | | Cite: | Crystal Structure of Glucokinase Regulatory Protein.
Biochemistry, 52, 2013
|
|
4BB9
 
 | | Crystal structure of glucokinase regulatory protein complexed to fructose-1-phosphate | | Descriptor: | 1-O-phosphono-beta-D-fructopyranose, CALCIUM ION, GLUCOKINASE REGULATORY PROTEIN | | Authors: | Pautsch, A, Stadler, N, Loehle, A, Lenter, M, Rist, W, Berg, A, Glocker, L, Nar, H, Reinert, D, Heckel, A, Schnapp, G, Kauschke, S.G. | | Deposit date: | 2012-09-21 | | Release date: | 2013-05-15 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (1.47 Å) | | Cite: | Crystal Structure of Glucokinase Regulatory Protein.
Biochemistry, 52, 2013
|
|
2A9K
 
 | | Crystal structure of the C3bot-NAD-RalA complex reveals a novel type of action of a bacterial exoenzyme | | Descriptor: | GUANOSINE-5'-DIPHOSPHATE, MAGNESIUM ION, Mono-ADP-ribosyltransferase C3, ... | | Authors: | Pautsch, A, Vogelsgesang, M, Trankle, J, Herrmann, C, Aktories, K. | | Deposit date: | 2005-07-12 | | Release date: | 2005-10-11 | | Last modified: | 2023-08-23 | | Method: | X-RAY DIFFRACTION (1.73 Å) | | Cite: | Crystal structure of the C3bot-RalA complex reveals a novel type of action of a bacterial exoenzyme.
Embo J., 24, 2005
|
|
2A78
 
 | | Crystal structure of the C3bot-RalA complex reveals a novel type of action of a bacterial exoenzyme | | Descriptor: | GUANOSINE-5'-DIPHOSPHATE, MAGNESIUM ION, Mono-ADP-ribosyltransferase C3, ... | | Authors: | Pautsch, A, Vogelsgesang, M, Trankle, J, Herrmann, C, Aktories, K. | | Deposit date: | 2005-07-05 | | Release date: | 2005-10-11 | | Last modified: | 2023-08-23 | | Method: | X-RAY DIFFRACTION (1.81 Å) | | Cite: | Crystal structure of the C3bot-RalA complex reveals a novel type of action of a bacterial exoenzyme.
Embo J., 24, 2005
|
|
1BXW
 
 | |
8OMJ
 
 | | hKHK-C in complex with compound 28 | | Descriptor: | Ketohexokinase, SULFATE ION, [3-[[6-[(3~{a}~{R},6~{a}~{S})-2,3,3~{a},4,6,6~{a}-hexahydro-1~{H}-pyrrolo[3,4-c]pyrrol-5-yl]-3-cyano-4-(trifluoromethyl)pyridin-2-yl]amino]-4-methylsulfanyl-phenyl]methoxy-methyl-phosphinic acid | | Authors: | Pautsch, A, Ebenhoch, R. | | Deposit date: | 2023-03-31 | | Release date: | 2024-07-10 | | Last modified: | 2024-09-04 | | Method: | X-RAY DIFFRACTION (1.978 Å) | | Cite: | Discovery of BI-9787, a potent zwitterionic ketohexokinase inhibitor with oral bioavailability.
Bioorg.Med.Chem.Lett., 112, 2024
|
|
6GPX
 
 | | CRYSTAL STRUCTURE OF CCR2A IN COMPLEX WITH MK-0812 | | Descriptor: | C-C chemokine receptor type 2,Rubredoxin,C-C chemokine receptor type 2, OLEIC ACID, ZINC ION, ... | | Authors: | Pautsch, A, Schnapp, G, Cheng, R, Apel, A. | | Deposit date: | 2018-06-07 | | Release date: | 2019-01-02 | | Last modified: | 2024-11-13 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | Crystal Structure of CC Chemokine Receptor 2A in Complex with an Orthosteric Antagonist Provides Insights for the Design of Selective Antagonists.
Structure, 27, 2019
|
|
6GPS
 
 | | CRYSTAL STRUCTURE OF CCR2A IN COMPLEX WITH MK-0812 | | Descriptor: | C-C chemokine receptor type 2,Rubredoxin,C-C chemokine receptor type 2, ZINC ION, [(3~{S},4~{S})-3-methoxyoxan-4-yl]-[(1~{R},3~{S})-3-propan-2-yl-3-[[3-(trifluoromethyl)-7,8-dihydro-5~{H}-1,6-naphthyridin-6-yl]carbonyl]cyclopentyl]azanium | | Authors: | Pautsch, A, Schnapp, G. | | Deposit date: | 2018-06-07 | | Release date: | 2019-01-02 | | Last modified: | 2024-11-20 | | Method: | X-RAY DIFFRACTION (3.3 Å) | | Cite: | Crystal Structure of CC Chemokine Receptor 2A in Complex with an Orthosteric Antagonist Provides Insights for the Design of Selective Antagonists.
Structure, 27, 2019
|
|
3P0G
 
 | | Structure of a nanobody-stabilized active state of the beta2 adrenoceptor | | Descriptor: | 8-[(1R)-2-{[1,1-dimethyl-2-(2-methylphenyl)ethyl]amino}-1-hydroxyethyl]-5-hydroxy-2H-1,4-benzoxazin-3(4H)-one, Beta-2 adrenergic receptor, Lysozyme, ... | | Authors: | Rasmussen, S.G.F, Choi, H.-J, Fung, J.J, Pardon, E, Casarosa, P, Chae, P.S, DeVree, B.T, Rosenbaum, D.M, Thian, F.S, Kobilka, T.S, Schnapp, A, Konetzki, I, Sunahara, R.K, Gellman, S.H, Pautsch, A, Steyaert, J, Weis, W.I, Kobilka, B.K. | | Deposit date: | 2010-09-28 | | Release date: | 2011-01-19 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (3.5 Å) | | Cite: | Structure of a nanobody-stabilized active state of the b2 adrenoceptor
Nature, 469, 2011
|
|
9FHE
 
 | | hKHK-C in complex with BI-9787 (pH 5.5) | | Descriptor: | (2~{S})-3-[3-[[4-[bis(fluoranyl)methyl]-3-cyano-6-[(3~{S})-3-(dimethylamino)pyrrolidin-1-yl]pyridin-2-yl]amino]-4-methylsulfanyl-phenyl]-2-methyl-propanoic acid, Ketohexokinase | | Authors: | Ebenhoch, R, Pautsch, A. | | Deposit date: | 2024-05-27 | | Release date: | 2024-09-04 | | Method: | X-RAY DIFFRACTION (2.313 Å) | | Cite: | Discovery of BI-9787, a potent zwitterionic ketohexokinase inhibitor with oral bioavailability.
Bioorg.Med.Chem.Lett., 112, 2024
|
|
9FHD
 
 | | hKHK-C in fomplex with BI-9787 | | Descriptor: | (2~{S})-3-[3-[[4-[bis(fluoranyl)methyl]-3-cyano-6-[(3~{S})-3-(dimethylamino)pyrrolidin-1-yl]pyridin-2-yl]amino]-4-methylsulfanyl-phenyl]-2-methyl-propanoic acid, Ketohexokinase, SULFATE ION | | Authors: | Ebenhoch, R, Pautsch, A. | | Deposit date: | 2024-05-27 | | Release date: | 2024-09-04 | | Method: | X-RAY DIFFRACTION (1.845 Å) | | Cite: | Discovery of BI-9787, a potent zwitterionic ketohexokinase inhibitor with oral bioavailability.
Bioorg.Med.Chem.Lett., 112, 2024
|
|
3BW8
 
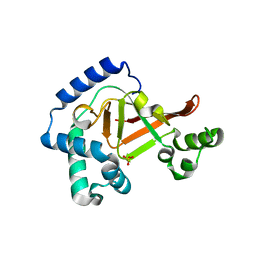 | | Crystal structure of the Clostridium limosum C3 exoenzyme | | Descriptor: | Mono-ADP-ribosyltransferase C3, SULFATE ION | | Authors: | Vogelsgesang, M, Stieglitz, B, Herrmann, C, Pautsch, A, Aktories, K. | | Deposit date: | 2008-01-08 | | Release date: | 2008-04-01 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Crystal structure of the Clostridium limosum C3 exoenzyme.
Febs Lett., 582, 2008
|
|
8QZ6
 
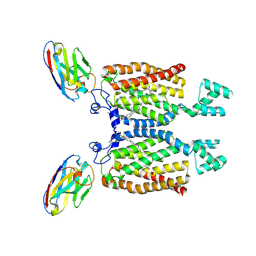 | | Structure of human ceramide synthase 6 (CerS6) bound to C16:0 | | Descriptor: | 1,2-DIACYL-SN-GLYCERO-3-PHOSPHOCHOLINE, 2-acetamido-2-deoxy-beta-D-glucopyranose, Isoform 2 of Ceramide synthase 6, ... | | Authors: | Pascoa, T.C, Pike, A.C.W, Chi, G, Stefanic, S, Quigley, A, Chalk, R, Mukhopadhyay, S.M.M, Venkaya, S, Dix, C, Moreira, T, Tessitore, A, Cole, V, Chu, A, Elkins, J.M, Pautsch, A, Schnapp, G, Carpenter, E.P, Sauer, D.B. | | Deposit date: | 2023-10-26 | | Release date: | 2024-11-13 | | Last modified: | 2025-03-26 | | Method: | ELECTRON MICROSCOPY (3.2 Å) | | Cite: | Structural basis of the mechanism and inhibition of a human ceramide synthase.
Nat.Struct.Mol.Biol., 32, 2025
|
|
8QZ7
 
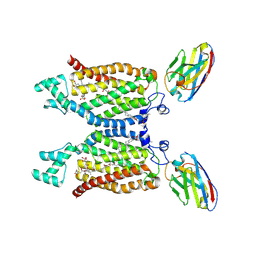 | | Structure of human ceramide synthase 6 (CerS6) in complex with N-palmitoyl fumonisin B1 | | Descriptor: | (2~{R})-2-[2-[(5~{R},6~{R},7~{S},9~{S},11~{R},16~{R},18~{S},19~{S})-6-[(3~{R})-3-carboxy-5-oxidanyl-5-oxidanylidene-pentanoyl]oxy-19-(hexadecanoylamino)-5,9-dimethyl-11,16,18-tris(oxidanyl)icosan-7-yl]oxy-2-oxidanylidene-ethyl]butanedioic acid, 1,2-DIACYL-SN-GLYCERO-3-PHOSPHOCHOLINE, 2-acetamido-2-deoxy-beta-D-glucopyranose, ... | | Authors: | Pascoa, T.C, Pike, A.C.W, Chi, G, Stefanic, S, Quigley, A, Chalk, R, Mukhopadhyay, S.M.M, Venkaya, S, Dix, C, Moreira, T, Tessitore, A, Cole, V, Chu, A, Elkins, J.M, Pautsch, A, Schnapp, G, Carpenter, E.P, Sauer, D.B. | | Deposit date: | 2023-10-26 | | Release date: | 2024-11-13 | | Last modified: | 2025-03-26 | | Method: | ELECTRON MICROSCOPY (3 Å) | | Cite: | Structural basis of the mechanism and inhibition of a human ceramide synthase.
Nat.Struct.Mol.Biol., 32, 2025
|
|
8OMK
 
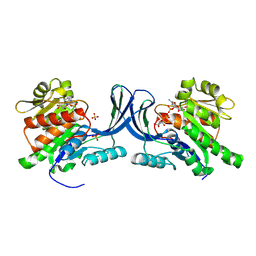 | | hKHK-C in complex with ADP & fructose 1-phosphate | | Descriptor: | 1-O-phosphono-beta-D-fructofuranose, ADENOSINE-5'-DIPHOSPHATE, Ketohexokinase, ... | | Authors: | Ebenhoch, R, Pautsch, A. | | Deposit date: | 2023-03-31 | | Release date: | 2024-01-10 | | Last modified: | 2024-09-04 | | Method: | X-RAY DIFFRACTION (2.48 Å) | | Cite: | Discovery of BI-9787, a potent zwitterionic ketohexokinase inhibitor with oral bioavailability.
Bioorg.Med.Chem.Lett., 112, 2024
|
|
8OME
 
 | | Crystal structure of hKHK-A in complex with compound-4 | | Descriptor: | Ketohexokinase, compound | | Authors: | Ebenhoch, R, Pautsch, A. | | Deposit date: | 2023-03-31 | | Release date: | 2023-09-27 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Crystal structures of human and mouse ketohexokinase provide a structural basis for species- and isoform-selective inhibitor design.
Acta Crystallogr D Struct Biol, 79, 2023
|
|
8OMD
 
 | | Crystal structure of mKHK in complex with compound-4 | | Descriptor: | Ketohexokinase, compound | | Authors: | Ebenhoch, R, Pautsch, A. | | Deposit date: | 2023-03-31 | | Release date: | 2023-09-27 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Crystal structures of human and mouse ketohexokinase provide a structural basis for species- and isoform-selective inhibitor design.
Acta Crystallogr D Struct Biol, 79, 2023
|
|
8OMF
 
 | | Crystal structure of hKHK-C in complex with compound-4 | | Descriptor: | Ketohexokinase, SULFATE ION, compound | | Authors: | Ebenhoch, R, Pautsch, A. | | Deposit date: | 2023-03-31 | | Release date: | 2023-09-27 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (2.14 Å) | | Cite: | Crystal structures of human and mouse ketohexokinase provide a structural basis for species- and isoform-selective inhibitor design.
Acta Crystallogr D Struct Biol, 79, 2023
|
|
9EOT
 
 | | Structure of human ceramide synthase 6 (CerS6) bound to C16:0 (nanobody Nb02) | | Descriptor: | 1,2-DIACYL-SN-GLYCERO-3-PHOSPHOCHOLINE, 2-acetamido-2-deoxy-beta-D-glucopyranose, Isoform 2 of Ceramide synthase 6, ... | | Authors: | Pascoa, T.C, Pike, A.C.W, Chi, G, Stefanic, S, Quigley, A, Chalk, R, Mukhopadhyay, S.M.M, Venkaya, S, Dix, C, Moreira, T, Tessitore, A, Cole, V, Chu, A, Elkins, J.M, Pautsch, A, Schnapp, G, Carpenter, E.P, Sauer, D.B. | | Deposit date: | 2024-03-15 | | Release date: | 2024-11-13 | | Last modified: | 2025-03-26 | | Method: | ELECTRON MICROSCOPY (3.02 Å) | | Cite: | Structural basis of the mechanism and inhibition of a human ceramide synthase.
Nat.Struct.Mol.Biol., 32, 2025
|
|
7QY1
 
 | | X-ray structure of furin in complex with the dichlorophenylpyridine-based inhibitor 4 | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 3-[4-[5-[4-[[4-(acetamidomethyl)piperidin-1-ium-1-yl]methyl]-6-[3,5-bis(chloranyl)phenyl]pyridin-2-yl]oxypyridin-2-yl]piperazin-1-ium-1-yl]propanoate, CALCIUM ION, ... | | Authors: | Dahms, S.O, Brandstetter, H, Pautsch, A. | | Deposit date: | 2022-01-27 | | Release date: | 2022-04-20 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (1.45 Å) | | Cite: | Dichlorophenylpyridine-Based Molecules Inhibit Furin through an Induced-Fit Mechanism.
Acs Chem.Biol., 17, 2022
|
|
7QXZ
 
 | | X-ray structure of furin in complex with the dichlorophenylpyridine-based inhibitor 5 | | Descriptor: | 2-[(3S)-1-[[2-[3,5-bis(chloranyl)phenyl]-6-[2-(4-methylpiperazin-4-ium-1-yl)pyrimidin-5-yl]oxy-pyridin-4-yl]methyl]pyrrolidin-1-ium-3-yl]oxyethanoic acid, CALCIUM ION, CHLORIDE ION, ... | | Authors: | Dahms, S.O, Brandstetter, H, Pautsch, A. | | Deposit date: | 2022-01-27 | | Release date: | 2022-04-13 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Dichlorophenylpyridine-Based Molecules Inhibit Furin through an Induced-Fit Mechanism.
Acs Chem.Biol., 17, 2022
|
|
7QY2
 
 | | X-ray structure of furin in complex with the dichlorophenylpyridine-based inhibitor 2 | | Descriptor: | (2R)-4-[4-[5-[4-[[4-(acetamidomethyl)piperidin-1-ium-1-yl]methyl]-6-[3,5-bis(chloranyl)phenyl]pyridin-2-yl]oxypyrimidin-2-yl]piperazin-1-ium-1-yl]-2-methyl-butanoate, 2-acetamido-2-deoxy-beta-D-glucopyranose, CALCIUM ION, ... | | Authors: | Dahms, S.O, Brandstetter, H, Pautsch, A. | | Deposit date: | 2022-01-27 | | Release date: | 2022-04-20 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (1.55 Å) | | Cite: | Dichlorophenylpyridine-Based Molecules Inhibit Furin through an Induced-Fit Mechanism.
Acs Chem.Biol., 17, 2022
|
|
