5Y63
 
 | | Crystal structure of Enterococcus faecalis AhpC | | Descriptor: | Alkyl hydroperoxide reductase, C subunit | | Authors: | Pan, A, Balakrishna, A.M, Grueber, G. | | Deposit date: | 2017-08-10 | | Release date: | 2017-12-27 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (2.87 Å) | | Cite: | Atomic structure and enzymatic insights into the vancomycin-resistant Enterococcus faecalis (V583) alkylhydroperoxide reductase subunit C
Free Radic. Biol. Med., 115, 2017
|
|
1D60
 
 | | THE STRUCTURE OF THE B-DNA DECAMER C-C-A-A-C-I-T-T-G-G: TRIGONAL FORM | | Descriptor: | DNA (5'-D(*CP*CP*AP*AP*CP*IP*TP*TP*GP*G)-3'), MAGNESIUM ION | | Authors: | Lipanov, A, Kopka, M.L, Kaczor-Grzeskowiak, M, Quintana, J, Dickerson, R.E. | | Deposit date: | 1992-02-26 | | Release date: | 1993-04-15 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Structure of the B-DNA decamer C-C-A-A-C-I-T-T-G-G in two different space groups: conformational flexibility of B-DNA.
Biochemistry, 32, 1993
|
|
1D61
 
 | | THE STRUCTURE OF THE B-DNA DECAMER C-C-A-A-C-I-T-T-G-G: MONOCLINIC FORM | | Descriptor: | CACODYLATE ION, CALCIUM ION, DNA (5'-D(*CP*CP*AP*AP*CP*IP*TP*TP*GP*G)-3') | | Authors: | Lipanov, A, Kopka, M.L, Kaczor-Grzeskowiak, M, Quintana, J, Dickerson, R.E. | | Deposit date: | 1992-02-26 | | Release date: | 1993-04-15 | | Last modified: | 2023-07-26 | | Method: | X-RAY DIFFRACTION (1.3 Å) | | Cite: | Structure of the B-DNA decamer C-C-A-A-C-I-T-T-G-G in two different space groups: conformational flexibility of B-DNA.
Biochemistry, 32, 1993
|
|
7P6R
 
 | | Crystal structure of the FimH-binding decoy module of human glycoprotein 2 (GP2) (crystal form I) | | Descriptor: | 1,2-ETHANEDIOL, 2-acetamido-2-deoxy-beta-D-glucopyranose, Isoform Alpha of Pancreatic secretory granule membrane major glycoprotein GP2 | | Authors: | Stsiapanava, A, Tunyasuvunakool, K, Jumper, J, de Sanctis, D, Jovine, L. | | Deposit date: | 2021-07-17 | | Release date: | 2022-03-16 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Structure of the decoy module of human glycoprotein 2 and uromodulin and its interaction with bacterial adhesin FimH.
Nat.Struct.Mol.Biol., 29, 2022
|
|
7P6S
 
 | | Crystal structure of the FimH-binding decoy module of human glycoprotein 2 (GP2) (crystal form II) | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, Isoform Alpha of Pancreatic secretory granule membrane major glycoprotein GP2, pentane-1,5-diol | | Authors: | Stsiapanava, A, Tunyasuvunakool, K, Jumper, J, de Sanctis, D, Jovine, L. | | Deposit date: | 2021-07-17 | | Release date: | 2022-03-16 | | Last modified: | 2024-11-13 | | Method: | X-RAY DIFFRACTION (1.35 Å) | | Cite: | Structure of the decoy module of human glycoprotein 2 and uromodulin and its interaction with bacterial adhesin FimH.
Nat.Struct.Mol.Biol., 29, 2022
|
|
7P6T
 
 | | Crystal structure of the FimH-binding decoy module of human glycoprotein 2 (GP2) (crystal form III) | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-ethyl-2-(hydroxymethyl)propane-1,3-diol, Isoform Alpha of Pancreatic secretory granule membrane major glycoprotein GP2 | | Authors: | Stsiapanava, A, Tunyasuvunakool, K, Jumper, J, de Sanctis, D, Jovine, L. | | Deposit date: | 2021-07-17 | | Release date: | 2022-03-16 | | Last modified: | 2024-10-09 | | Method: | X-RAY DIFFRACTION (1.4 Å) | | Cite: | Structure of the decoy module of human glycoprotein 2 and uromodulin and its interaction with bacterial adhesin FimH.
Nat.Struct.Mol.Biol., 29, 2022
|
|
6TQK
 
 | | Cryo-EM of native human uromodulin (UMOD)/Tamm-Horsfall protein (THP) filament. | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Uromodulin, ... | | Authors: | Stsiapanava, A, Xu, C, Carroni, M, Wu, B, Jovine, L. | | Deposit date: | 2019-12-16 | | Release date: | 2020-11-04 | | Last modified: | 2024-10-09 | | Method: | ELECTRON MICROSCOPY (3.35 Å) | | Cite: | Cryo-EM structure of native human uromodulin, a zona pellucida module polymer.
Embo J., 39, 2020
|
|
6TQL
 
 | | Cryo-EM of elastase-treated human uromodulin (UMOD)/Tamm-Horsfall protein (THP) filament | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-2)-[2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)]alpha-D-mannopyranose-(1-3)-[2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-alpha-D-mannopyranose-(1-6)]beta-D-mannopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, ... | | Authors: | Stsiapanava, A, Xu, C, Carroni, M, Wu, B, Jovine, L. | | Deposit date: | 2019-12-16 | | Release date: | 2020-11-04 | | Last modified: | 2024-11-13 | | Method: | ELECTRON MICROSCOPY (3.96 Å) | | Cite: | Cryo-EM structure of native human uromodulin, a zona pellucida module polymer.
Embo J., 39, 2020
|
|
6NVM
 
 | |
2GVF
 
 | | HCV NS3-4A protease domain complexed with a macrocyclic ketoamide inhibitor, SCH419021 | | Descriptor: | (6R,8S,11S)-11-CYCLOHEXYL-N-(1-{[(2-{[(1S)-2-(DIMETHYLAMINO)-2-OXO-1-PHENYLETHYL]AMINO}-2-OXOETHYL)AMINO](OXO)ACETYL}BUTYL)-10,13-DIOXO-2,5-DIOXA-9,12-DIAZATRICYCLO[14.3.1.1~6,9~]HENICOSA-1(20),16,18-TRIENE-8-CARBOXAMIDE, Polyprotein, ZINC ION, ... | | Authors: | Arasappan, A, Njoroge, F.G, Chen, K.X, Venkatraman, S, Parekh, T.N, Gu, H, Pichardo, J, Butkiewicz, N, Prongay, A, Madison, V, Girijavallabhan, V. | | Deposit date: | 2006-05-02 | | Release date: | 2007-01-23 | | Last modified: | 2011-07-13 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | P2-P4 macrocyclic inhibitors of hepatitis C virus NS3-4A serine protease.
Bioorg.Med.Chem.Lett., 16, 2006
|
|
2I9F
 
 | | Structure of the equine arterivirus nucleocapsid protein | | Descriptor: | 2,3-DIHYDROXY-1,4-DITHIOBUTANE, CHLORIDE ION, GLYCEROL, ... | | Authors: | Deshpande, A, Wang, S, Walsh, M, Dokland, T. | | Deposit date: | 2006-09-05 | | Release date: | 2007-05-01 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Structure of the equine arteritis virus nucleocapsid protein reveals a dimer-dimer arrangement.
Acta Crystallogr.,Sect.D, 63, 2007
|
|
3RVA
 
 | | Crystal structure of organophosphorus acid anhydrolase from Alteromonas macleodii | | Descriptor: | MANGANESE (II) ION, NICKEL (II) ION, Organophosphorus acid anhydrolase, ... | | Authors: | Stepankova, A, Koval, T, Ostergaard, L.H, Duskova, J, Skalova, T, Hasek, J, Dohnalek, J. | | Deposit date: | 2011-05-06 | | Release date: | 2012-05-09 | | Last modified: | 2023-12-06 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Organophosphorus acid anhydrolase from Alteromonas macleodii: structural study and functional relationship to prolidases.
Acta Crystallogr.,Sect.F, 69, 2013
|
|
4MKT
 
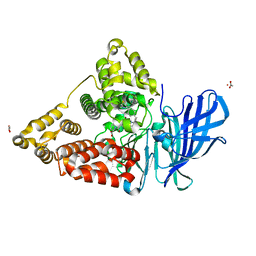 | | Human Leukotriene A4 Hydrolase in complex with Pro-Gly-Pro analogue and 4-(4-benzylphenyl)thiazol-2-amine | | Descriptor: | 1-{4-oxo-4-[(2S)-pyrrolidin-2-yl]butanoyl}-L-proline, 4-(4-benzylphenyl)-1,3-thiazol-2-amine, ACETIC ACID, ... | | Authors: | Stsiapanava, A, Rinaldo-Matthis, A, Haeggstrom, J.Z. | | Deposit date: | 2013-09-05 | | Release date: | 2014-03-12 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (1.618 Å) | | Cite: | Binding of Pro-Gly-Pro at the active site of leukotriene A4 hydrolase/aminopeptidase and development of an epoxide hydrolase selective inhibitor.
Proc.Natl.Acad.Sci.USA, 111, 2014
|
|
4L2L
 
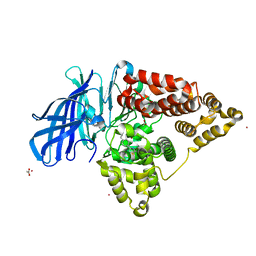 | | Human Leukotriene A4 Hydrolase complexed with ligand 4-(4-benzylphenyl)thiazol-2-amine | | Descriptor: | 4-(4-benzylphenyl)-1,3-thiazol-2-amine, ACETATE ION, Leukotriene A-4 hydrolase, ... | | Authors: | Stsiapanava, A, Rinaldo-Matthis, A, Haeggstrom, J.Z. | | Deposit date: | 2013-06-04 | | Release date: | 2014-03-12 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (1.648 Å) | | Cite: | Binding of Pro-Gly-Pro at the active site of leukotriene A4 hydrolase/aminopeptidase and development of an epoxide hydrolase selective inhibitor.
Proc.Natl.Acad.Sci.USA, 111, 2014
|
|
4MS6
 
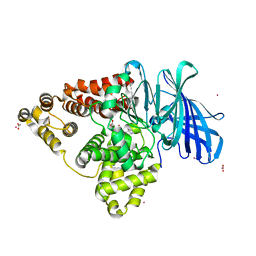 | | Human Leukotriene A4 Hydrolase in complex with Pro-Gly-Pro analogue | | Descriptor: | 1-{4-oxo-4-[(2S)-pyrrolidin-2-yl]butanoyl}-L-proline, ACETIC ACID, Leukotriene A-4 hydrolase, ... | | Authors: | Stsiapanava, A, Rinaldo-Matthis, A, Haeggstrom, J.Z. | | Deposit date: | 2013-09-18 | | Release date: | 2014-03-12 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (1.72 Å) | | Cite: | Binding of Pro-Gly-Pro at the active site of leukotriene A4 hydrolase/aminopeptidase and development of an epoxide hydrolase selective inhibitor.
Proc.Natl.Acad.Sci.USA, 111, 2014
|
|
4GAA
 
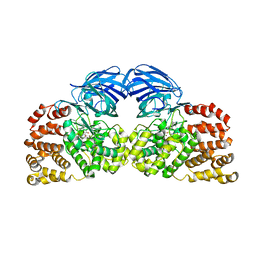 | | Structure of Leukotriene A4 hydrolase from Xenopus laevis complexed with inhibitor bestatin | | Descriptor: | 2-(3-AMINO-2-HYDROXY-4-PHENYL-BUTYRYLAMINO)-4-METHYL-PENTANOIC ACID, MGC78867 protein, ZINC ION | | Authors: | Stsiapanava, A, Kumar, R.B, Haeggstrom, J.Z, Rinaldo-Matthis, A. | | Deposit date: | 2012-07-25 | | Release date: | 2013-06-12 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (2.26 Å) | | Cite: | Product formation controlled by substrate dynamics in leukotriene A4 hydrolase.
Biochim.Biophys.Acta, 1844, 2014
|
|
8FAX
 
 | | Fab 1249A8-MERS Stem Helix Complex | | Descriptor: | 1249A8-HC, 1249A8-LC, CHLORIDE ION, ... | | Authors: | Deshpande, A, Schormann, N, Piepenbrink, M.S, Martinez-Sobrido, L, Kobie, J.J, Walter, M.R. | | Deposit date: | 2022-11-28 | | Release date: | 2023-05-03 | | Last modified: | 2023-07-19 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Structure and epitope of a neutralizing monoclonal antibody that targets the stem helix of beta coronaviruses.
Febs J., 290, 2023
|
|
4P7V
 
 | | Structural insights into higher-order assembly and function of the bacterial microcompartment protein PduA | | Descriptor: | GLYCEROL, Polyhedral bodies | | Authors: | Pang, A, Frank, S, Brown, I.R, Warren, M.J, Pickersgill, R.W. | | Deposit date: | 2014-03-27 | | Release date: | 2014-06-04 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (1.93 Å) | | Cite: | Structural Insights into Higher Order Assembly and Function of the Bacterial Microcompartment Protein PduA.
J.Biol.Chem., 289, 2014
|
|
1YKX
 
 | | Effect of alcohols on protein hydration | | Descriptor: | CHLORIDE ION, ETHANOL, Lysozyme C, ... | | Authors: | Deshpande, A, Nimsadkar, S, Mande, S.C. | | Deposit date: | 2005-01-18 | | Release date: | 2005-07-05 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Effect of alcohols on protein hydration: crystallographic analysis of hen egg-white lysozyme in the presence of alcohols.
Acta Crystallogr.,Sect.D, 61, 2005
|
|
1YKY
 
 | | Effect of alcohols on protein hydration | | Descriptor: | 1-BUTANOL, CHLORIDE ION, Lysozyme C, ... | | Authors: | Deshpande, A, Nimsadkar, S, Mande, S.C. | | Deposit date: | 2005-01-18 | | Release date: | 2005-07-05 | | Last modified: | 2024-11-13 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Effect of alcohols on protein hydration: crystallographic analysis of hen egg-white lysozyme in the presence of alcohols.
Acta Crystallogr.,Sect.D, 61, 2005
|
|
1YKZ
 
 | | Effect of alcohols on protein hydration | | Descriptor: | CHLORIDE ION, Lysozyme C, PENTANAL, ... | | Authors: | Deshpande, A, Nimsadkar, S, Mande, S.C. | | Deposit date: | 2005-01-18 | | Release date: | 2005-07-05 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Effect of alcohols on protein hydration: crystallographic analysis of hen egg-white lysozyme in the presence of alcohols.
Acta Crystallogr.,Sect.D, 61, 2005
|
|
1YL0
 
 | | Effect of alcohols on protein hydration | | Descriptor: | CHLORIDE ION, ISOPROPYL ALCOHOL, Lysozyme C, ... | | Authors: | Deshpande, A, Nimsadkar, S, Mande, S.C. | | Deposit date: | 2005-01-18 | | Release date: | 2005-07-05 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Effect of alcohols on protein hydration: crystallographic analysis of hen egg-white lysozyme in the presence of alcohols.
Acta Crystallogr.,Sect.D, 61, 2005
|
|
1Z55
 
 | | Effect of alcohols on protein hydration | | Descriptor: | CHLORIDE ION, Lysozyme C, SODIUM ION | | Authors: | Deshpande, A, Nimsadkar, S, Mande, S.C. | | Deposit date: | 2005-03-17 | | Release date: | 2005-07-05 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Effect of alcohols on protein hydration: crystallographic analysis of hen egg-white lysozyme in the presence of alcohols.
Acta Crystallogr.,Sect.D, 61, 2005
|
|
2A4G
 
 | | Hepatitis C Protease NS3-4A serine protease with Ketoamide Inhibitor SCH225724 Bound | | Descriptor: | ({1-[1-CARBAMOYL-PHENYL-METHYL)-CARBAMOYL]-METHYL}-AMINOOXALYL)-BUTYLCARBAMOYL)-3-METHYL-BUTYLCARBAMOYL)-CYCLOHEXYL-METHYL)-CARBAMIC ACID ISOBUTYL ESTER, NS3 protease/helicase, NS4a peptide, ... | | Authors: | Arasappan, A, Njoroge, F.G, Chan, T.Y, Bennett, F, Bogen, S.L, Chen, K, Gu, H, Hong, L, Jao, E, Liu, Y.T, Lovey, R.G, Parekh, T, Pike, R.E, Pinto, P, Santhanam, B, Venkatraman, S, Vaccaro, H, Wang, H, Yang, X, Zhu, Z, Mckittrick, B, Saksena, A.K, Girijavallabhan, V, Pichardo, J, Butkiewicz, N, Ingram, R, Malcolm, B, Prongay, A.J, Yao, N, Marten, B, Madison, V, Kemp, S, Levy, O, Lim-Wilby, M, Tamura, S, Ganguly, A.K. | | Deposit date: | 2005-06-28 | | Release date: | 2006-07-04 | | Last modified: | 2024-10-09 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Hepatitis C virus NS3-4a serine protease inhibitors. SAR of P2' moiety with improved potency.
Bioorg.Med.Chem.Lett., 15, 2005
|
|
5NIA
 
 | |
