5OAM
 
 | | Molecular basis of human kinesin-8 function and inhibition | | Descriptor: | GUANOSINE-5'-DIPHOSPHATE, GUANOSINE-5'-TRIPHOSPHATE, Kinesin-like protein KIF18A, ... | | Authors: | Locke, J, Joseph, A.P, Topf, M, Moores, C.A. | | Deposit date: | 2017-06-23 | | Release date: | 2017-10-25 | | Last modified: | 2024-05-15 | | Method: | ELECTRON MICROSCOPY (5.5 Å) | | Cite: | Structural basis of human kinesin-8 function and inhibition.
Proc. Natl. Acad. Sci. U.S.A., 114, 2017
|
|
5OCU
 
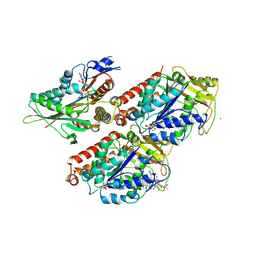 | | Molecular basis of human kinesin-8 function and inhibition | | Descriptor: | GUANOSINE-5'-DIPHOSPHATE, GUANOSINE-5'-TRIPHOSPHATE, Kinesin-like protein KIF18A, ... | | Authors: | Locke, J, Joseph, A.P, Topf, M, Moores, C.A. | | Deposit date: | 2017-07-03 | | Release date: | 2017-10-25 | | Last modified: | 2024-05-15 | | Method: | ELECTRON MICROSCOPY (5.2 Å) | | Cite: | Structural basis of human kinesin-8 function and inhibition.
Proc. Natl. Acad. Sci. U.S.A., 114, 2017
|
|
5OGC
 
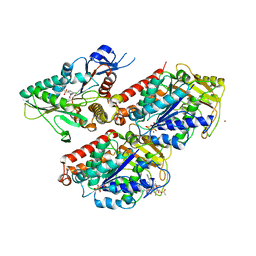 | | Molecular basis of human kinesin-8 function and inhibition | | Descriptor: | 4-chloranyl-2-nitro-1-(phenylsulfonyl)benzene, GUANOSINE-5'-DIPHOSPHATE, GUANOSINE-5'-TRIPHOSPHATE, ... | | Authors: | Locke, J, Joseph, A.P, Topf, M, Moores, C.A. | | Deposit date: | 2017-07-12 | | Release date: | 2017-10-25 | | Last modified: | 2024-05-15 | | Method: | ELECTRON MICROSCOPY (4.8 Å) | | Cite: | Structural basis of human kinesin-8 function and inhibition.
Proc. Natl. Acad. Sci. U.S.A., 114, 2017
|
|
6F0L
 
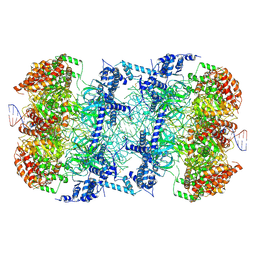 | | S. cerevisiae MCM double hexamer bound to duplex DNA | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, DNA (62-MER), DNA replication licensing factor MCM2, ... | | Authors: | Abid Ali, F, Pye, V.E, Douglas, M.E, Locke, J, Nans, A, Diffley, J.F.X, Costa, A. | | Deposit date: | 2017-11-20 | | Release date: | 2017-12-06 | | Last modified: | 2019-12-11 | | Method: | ELECTRON MICROSCOPY (4.77 Å) | | Cite: | Cryo-EM structure of a licensed DNA replication origin.
Nat Commun, 8, 2017
|
|
6ZPG
 
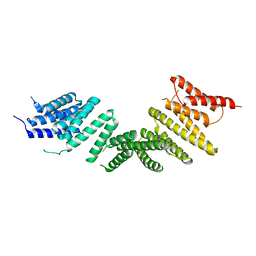 | | Kinesin binding protein (KBP) | | Descriptor: | KIF-binding protein | | Authors: | Atherton, J, Hummel, J.J.A, Olieric, N, Locke, J, Pena, A, Rosenfeld, S.S, Steinmetz, M.O, Hoogenraad, C.C, Moores, C.A. | | Deposit date: | 2020-07-08 | | Release date: | 2020-12-30 | | Last modified: | 2024-05-01 | | Method: | ELECTRON MICROSCOPY (4.6 Å) | | Cite: | The mechanism of kinesin inhibition by kinesin-binding protein.
Elife, 9, 2020
|
|
6ZPH
 
 | | Kinesin binding protein complexed with Kif15 motor domain | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, KIF-binding protein, Kinesin-like protein KIF15, ... | | Authors: | Atherton, J, Hummel, J.J.A, Olieric, N, Locke, J, Pena, A, Rosenfeld, S.S, Steinmetz, M.O, Hoogenraad, C.C, Moores, C.A. | | Deposit date: | 2020-07-08 | | Release date: | 2020-12-30 | | Last modified: | 2024-05-01 | | Method: | ELECTRON MICROSCOPY (6.9 Å) | | Cite: | The mechanism of kinesin inhibition by kinesin-binding protein.
Elife, 9, 2020
|
|
6ZPI
 
 | | Microtubule complexed with Kif15 motor domain. Symmetrised asymmetric unit | | Descriptor: | GUANOSINE-5'-DIPHOSPHATE, GUANOSINE-5'-TRIPHOSPHATE, Kinesin-like protein KIF15, ... | | Authors: | Atherton, J, Hummel, J.J.A, Olieric, N, Locke, J, Pena, A, Rosenfeld, S.S, Steinmetz, M.O, Hoogenraad, C.C, Moores, C.A. | | Deposit date: | 2020-07-08 | | Release date: | 2020-12-30 | | Last modified: | 2024-11-06 | | Method: | ELECTRON MICROSCOPY (4.5 Å) | | Cite: | The mechanism of kinesin inhibition by kinesin-binding protein.
Elife, 9, 2020
|
|
7ZPP
 
 | | Cryo-EM structure of the MVV CSC intasome at 4.5A resolution | | Descriptor: | Integrase, vDNA, non-transferred strand, ... | | Authors: | Ballandras-Colas, A, Maskell, D, Pye, V.E, Locke, J, Swuec, S, Kotecha, A, Costa, A, Cherepanov, P. | | Deposit date: | 2022-04-28 | | Release date: | 2022-05-11 | | Last modified: | 2024-07-24 | | Method: | ELECTRON MICROSCOPY (4.5 Å) | | Cite: | A supramolecular assembly mediates lentiviral DNA integration
Science, 355, 2017
|
|
5M0R
 
 | | Cryo-EM reconstruction of the maedi-visna virus (MVV) strand transfer complex | | Descriptor: | integrase, tDNA, vDNA, ... | | Authors: | Pye, V.E, Ballandras-Colas, A, Maskell, D, Locke, J, Kotecha, A, Costa, A, Cherepanov, P. | | Deposit date: | 2016-10-05 | | Release date: | 2017-01-18 | | Last modified: | 2024-05-15 | | Method: | ELECTRON MICROSCOPY (8.2 Å) | | Cite: | A supramolecular assembly mediates lentiviral DNA integration.
Science, 355, 2017
|
|
2JOZ
 
 | | Solution NMR structure of protein yxeF, Northeast Structural Genomics Consortium target Sr500a | | Descriptor: | Hypothetical protein yxeF | | Authors: | Wu, Y, Liu, G, Zhang, Q, Bhatnagar, S, Chen, C, Nwosu, C, Xiao, R, Cunningham, K, Locke, J, Ma, L, Swapna, G, Baran, M, Acton, T, Montelione, G, Szyperski, T, Northeast Structural Genomics Consortium (NESG) | | Deposit date: | 2007-04-10 | | Release date: | 2007-05-15 | | Last modified: | 2024-05-08 | | Method: | SOLUTION NMR | | Cite: | Solution NMR structure of protein yxeF , Northeast Structural Genomics Consortium target Sr500a
To be Published
|
|
6RQC
 
 | | Cryo-EM structure of an MCM loading intermediate | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, ADENOSINE-5'-TRIPHOSPHATE, DNA (88-MER), ... | | Authors: | Miller, T.C.R, Locke, J, Costa, A. | | Deposit date: | 2019-05-15 | | Release date: | 2019-11-20 | | Last modified: | 2024-05-22 | | Method: | ELECTRON MICROSCOPY (4.4 Å) | | Cite: | Mechanism of head-to-head MCM double-hexamer formation revealed by cryo-EM.
Nature, 575, 2019
|
|
5LLJ
 
 | |
7P30
 
 | | 3.0 A resolution structure of a DNA-loaded MCM double hexamer | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, ADENOSINE-5'-TRIPHOSPHATE, DNA (53-MER), ... | | Authors: | Greiwe, J.F, Miller, T.C.R, Martino, F, Costa, A. | | Deposit date: | 2021-07-06 | | Release date: | 2022-02-02 | | Last modified: | 2024-07-17 | | Method: | ELECTRON MICROSCOPY (3 Å) | | Cite: | Structural mechanism for the selective phosphorylation of DNA-loaded MCM double hexamers by the Dbf4-dependent kinase.
Nat.Struct.Mol.Biol., 29, 2022
|
|
7P5Z
 
 | | Structure of a DNA-loaded MCM double hexamer engaged with the Dbf4-dependent kinase | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, ADENOSINE-5'-TRIPHOSPHATE, Cell division control protein 7, ... | | Authors: | Greiwe, J.F, Miller, T.C.R, Martino, F, Costa, A. | | Deposit date: | 2021-07-15 | | Release date: | 2022-02-02 | | Last modified: | 2024-07-17 | | Method: | ELECTRON MICROSCOPY (3.3 Å) | | Cite: | Structural mechanism for the selective phosphorylation of DNA-loaded MCM double hexamers by the Dbf4-dependent kinase.
Nat.Struct.Mol.Biol., 29, 2022
|
|
6YUF
 
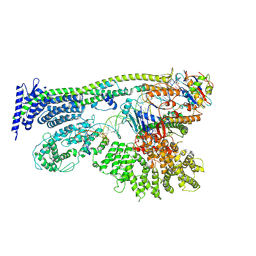 | | Cohesin complex with loader gripping DNA | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, BERYLLIUM TRIFLUORIDE ION, Cohesin subunit rad21, ... | | Authors: | Higashi, T.L, Eickhoff, P, Sousa, J.S, Costa, A, Uhlmann, F. | | Deposit date: | 2020-04-27 | | Release date: | 2020-08-19 | | Last modified: | 2024-05-22 | | Method: | ELECTRON MICROSCOPY (3.94 Å) | | Cite: | A Structure-Based Mechanism for DNA Entry into the Cohesin Ring.
Mol.Cell, 79, 2020
|
|
5T3A
 
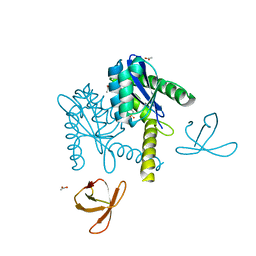 | |
6HV9
 
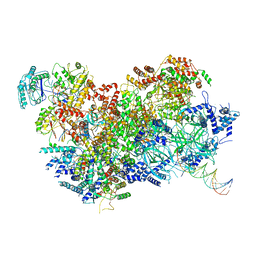 | | S. cerevisiae CMG-Pol epsilon-DNA | | Descriptor: | Cell division control protein 45, DNA (5'-D(*GP*CP*AP*GP*CP*CP*AP*CP*GP*CP*TP*GP*GP*CP*CP*GP*TP*TP*TP*TP*A)-3'), DNA (5'-D(P*TP*AP*AP*AP*AP*CP*GP*GP*CP*CP*AP*GP*CP*GP*TP*GP*GP*CP*TP*GP*C)-3'), ... | | Authors: | Abid Ali, F, Purkiss, A.G, Cheung, A, Costa, A. | | Deposit date: | 2018-10-10 | | Release date: | 2018-12-12 | | Last modified: | 2023-07-05 | | Method: | ELECTRON MICROSCOPY (4.98 Å) | | Cite: | Structure of DNA-CMG-Pol epsilon elucidates the roles of the non-catalytic polymerase modules in the eukaryotic replisome.
Nat Commun, 9, 2018
|
|
6HV8
 
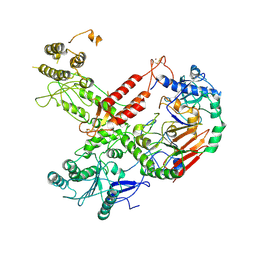 | | Cryo-EM structure of S. cerevisiae Polymerase epsilon deltacat mutant | | Descriptor: | DNA polymerase epsilon catalytic subunit A, DNA polymerase epsilon subunit B, ZINC ION | | Authors: | Goswami, P, Purkiss, A, Cheung, A, Costa, A. | | Deposit date: | 2018-10-10 | | Release date: | 2018-12-12 | | Last modified: | 2024-10-16 | | Method: | ELECTRON MICROSCOPY (4.4 Å) | | Cite: | Structure of DNA-CMG-Pol epsilon elucidates the roles of the non-catalytic polymerase modules in the eukaryotic replisome.
Nat Commun, 9, 2018
|
|
2K1G
 
 | | Solution NMR structure of lipoprotein spr from Escherichia coli K12. Northeast Structural Genomics target ER541-37-162 | | Descriptor: | Lipoprotein spr | | Authors: | Aramini, J.M, Rossi, P, Zhao, L, Jiang, M, Maglaqui, M, Xiao, R, Liu, J, Baran, M.C, Swapna, G.V.T, Huang, Y.J, Acton, T.B, Rost, B, Montelione, G.T, Northeast Structural Genomics Consortium (NESG) | | Deposit date: | 2008-03-03 | | Release date: | 2008-03-18 | | Last modified: | 2024-05-01 | | Method: | SOLUTION NMR | | Cite: | Solution NMR structure of the NlpC/P60 domain of lipoprotein Spr from Escherichia coli: structural evidence for a novel cysteine peptidase catalytic triad.
Biochemistry, 47, 2008
|
|
6RAZ
 
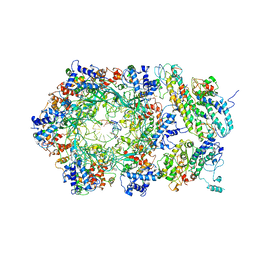 | | D. melanogaster CMG-DNA, State 2B | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, ADENOSINE-5'-TRIPHOSPHATE, AT18545p, ... | | Authors: | Eickhoff, P, Martino, F, Costa, A. | | Deposit date: | 2019-04-08 | | Release date: | 2019-09-18 | | Last modified: | 2021-01-20 | | Method: | ELECTRON MICROSCOPY (4.46 Å) | | Cite: | Molecular Basis for ATP-Hydrolysis-Driven DNA Translocation by the CMG Helicase of the Eukaryotic Replisome.
Cell Rep, 28, 2019
|
|
6RAY
 
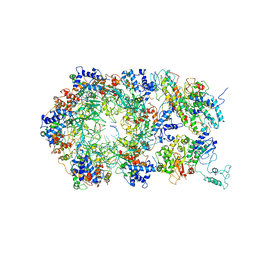 | | D. melanogaster CMG-DNA, State 2A | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, ADENOSINE-5'-TRIPHOSPHATE, AT18545p, ... | | Authors: | Eickhoff, P, Martino, F, Costa, A. | | Deposit date: | 2019-04-08 | | Release date: | 2019-09-11 | | Last modified: | 2024-10-23 | | Method: | ELECTRON MICROSCOPY (4.28 Å) | | Cite: | Molecular Basis for ATP-Hydrolysis-Driven DNA Translocation by the CMG Helicase of the Eukaryotic Replisome.
Cell Rep, 28, 2019
|
|
6RAX
 
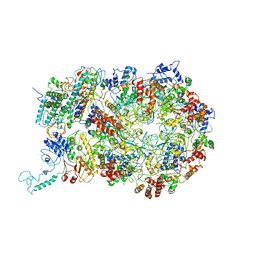 | | D. melanogaster CMG-DNA, State 1B | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, ADENOSINE-5'-TRIPHOSPHATE, AT18545p, ... | | Authors: | Eickhoff, P, Martino, F, Costa, A. | | Deposit date: | 2019-04-08 | | Release date: | 2019-09-11 | | Last modified: | 2019-09-18 | | Method: | ELECTRON MICROSCOPY (3.99 Å) | | Cite: | Molecular Basis for ATP-Hydrolysis-Driven DNA Translocation by the CMG Helicase of the Eukaryotic Replisome.
Cell Rep, 28, 2019
|
|
6RAW
 
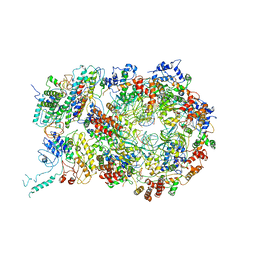 | | D. melanogaster CMG-DNA, State 1A | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, ADENOSINE-5'-TRIPHOSPHATE, AT18545p, ... | | Authors: | Eickhoff, P, Martino, F, Costa, A. | | Deposit date: | 2019-04-08 | | Release date: | 2019-09-11 | | Last modified: | 2024-10-16 | | Method: | ELECTRON MICROSCOPY (3.7 Å) | | Cite: | Molecular Basis for ATP-Hydrolysis-Driven DNA Translocation by the CMG Helicase of the Eukaryotic Replisome.
Cell Rep, 28, 2019
|
|
6ZH0
 
 | | Structure of human galactokinase 1 bound with 2-(4-chlorophenyl)-N-(pyrimidin-2-yl)acetamide | | Descriptor: | 2-(1,3-benzoxazol-2-ylamino)spiro[1,6,7,8-tetrahydroquinazoline-4,1'-cyclohexane]-5-one, Galactokinase, N-(3-chlorophenyl)-2,2,2-trifluoroacetamide, ... | | Authors: | Mackinnon, S.R, Bezerra, G.A, Zhang, M, Foster, W, Krojer, T, Brandao-Neto, J, Douangamath, A, Arrowsmith, C, Edwards, A, Bountra, C, Brennan, P, Lai, K, Yue, W.W. | | Deposit date: | 2020-06-20 | | Release date: | 2022-06-01 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Fragment Screening Reveals Starting Points for Rational Design of Galactokinase 1 Inhibitors to Treat Classic Galactosemia.
Acs Chem.Biol., 16, 2021
|
|
2Z9I
 
 | |
