7S13
 
 | | Crystal structure of Fab in complex with mouse CD96 dimer | | Descriptor: | 1,2-ETHANEDIOL, 2-acetamido-2-deoxy-beta-D-glucopyranose, CITRATE ANION, ... | | Authors: | Lee, P.S, Barman, I, Strop, P. | | Deposit date: | 2021-08-31 | | Release date: | 2021-10-20 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (2.12 Å) | | Cite: | Antibody blockade of CD96 by distinct molecular mechanisms.
Mabs, 13, 2021
|
|
7S11
 
 | | Crystal structure of Fab in complex with mouse CD96 monomer | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, Fab heavy chain, Fab light chain, ... | | Authors: | Lee, P.S, Chau, B, Strop, P. | | Deposit date: | 2021-08-31 | | Release date: | 2021-11-03 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (2.58 Å) | | Cite: | Antibody blockade of CD96 by distinct molecular mechanisms.
Mabs, 13, 2021
|
|
4WUK
 
 | | Crystal structure of apo CH65 Fab | | Descriptor: | 2-[N-CYCLOHEXYLAMINO]ETHANE SULFONIC ACID, CH65 heavy chain, CH65 light chain | | Authors: | Lee, P.S, Wilson, I.A. | | Deposit date: | 2014-11-01 | | Release date: | 2015-02-25 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Structure of the apo anti-influenza CH65 Fab.
Acta Crystallogr.,Sect.F, 71, 2015
|
|
4ZCJ
 
 | |
4GMT
 
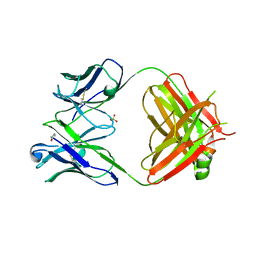 | | Crystal structure of heterosubtypic Fab S139/1 | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Fab S139/1 heavy chain, ... | | Authors: | Lee, P.S, Ekiert, D.C, Wilson, I.A. | | Deposit date: | 2012-08-16 | | Release date: | 2012-10-03 | | Last modified: | 2024-10-09 | | Method: | X-RAY DIFFRACTION (2.05 Å) | | Cite: | Heterosubtypic antibody recognition of the influenza virus hemagglutinin receptor binding site enhanced by avidity.
Proc.Natl.Acad.Sci.USA, 109, 2012
|
|
7SU1
 
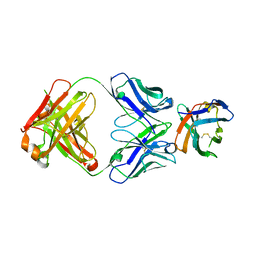 | | Crystal structure of an acidic pH-selective Ipilimumab variant Ipi.106 in complex with CTLA-4 | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, Cytotoxic T-lymphocyte protein 4, Fab heavy chain, ... | | Authors: | Lee, P.S, Chau, B, Strop, P. | | Deposit date: | 2021-11-15 | | Release date: | 2022-03-02 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (2.53 Å) | | Cite: | Improved therapeutic index of an acidic pH-selective antibody.
Mabs, 14, 2022
|
|
7SU0
 
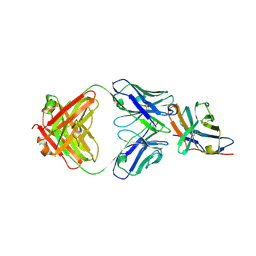 | | Crystal structure of an acidic pH-selective Ipilimumab variant Ipi.105 in complex with CTLA-4 | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, CITRATE ANION, Cytotoxic T-lymphocyte protein 4, ... | | Authors: | Lee, P.S, Chau, B, Strop, P. | | Deposit date: | 2021-11-15 | | Release date: | 2022-03-02 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (2.41 Å) | | Cite: | Improved therapeutic index of an acidic pH-selective antibody.
Mabs, 14, 2022
|
|
4GMS
 
 | | Crystal structure of heterosubtypic Fab S139/1 in complex with influenza A H3 hemagglutinin | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Fab S139/1 heavy chain, ... | | Authors: | Lee, P.S, Ekiert, D.C, Wilson, I.A. | | Deposit date: | 2012-08-16 | | Release date: | 2012-10-03 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (2.95 Å) | | Cite: | Heterosubtypic antibody recognition of the influenza virus hemagglutinin receptor binding site enhanced by avidity.
Proc.Natl.Acad.Sci.USA, 109, 2012
|
|
4O5I
 
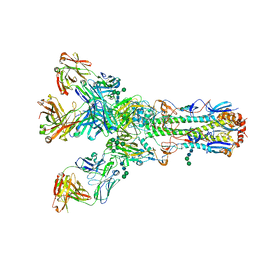 | |
4O5N
 
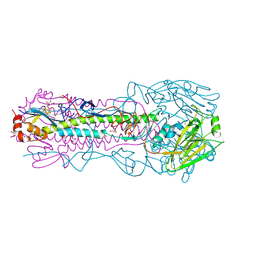 | | Crystal structure of A/Victoria/361/2011 (H3N2) influenza virus hemagglutinin | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, DI(HYDROXYETHYL)ETHER, ... | | Authors: | Lee, P.S, Wilson, I.A. | | Deposit date: | 2013-12-19 | | Release date: | 2014-04-16 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (1.75 Å) | | Cite: | Receptor mimicry by antibody F045-092 facilitates universal binding to the H3 subtype of influenza virus.
Nat Commun, 5, 2014
|
|
4O5L
 
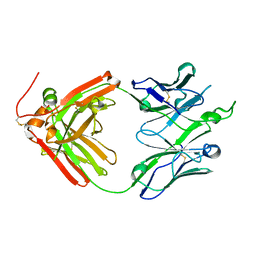 | | Crystal structure of broadly neutralizing antibody F045-092 | | Descriptor: | Fab F045-092 heavy chain, Fab F045-092 light chain, PHOSPHATE ION | | Authors: | Lee, P.S, Wilson, I.A. | | Deposit date: | 2013-12-19 | | Release date: | 2014-04-16 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (1.5045 Å) | | Cite: | Receptor mimicry by antibody F045-092 facilitates universal binding to the H3 subtype of influenza virus.
Nat Commun, 5, 2014
|
|
4O58
 
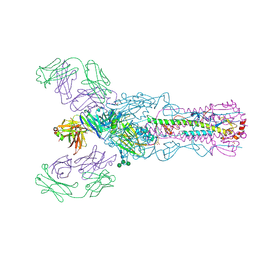 | |
1X3P
 
 | |
5KUX
 
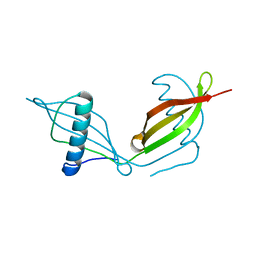 | |
7S7I
 
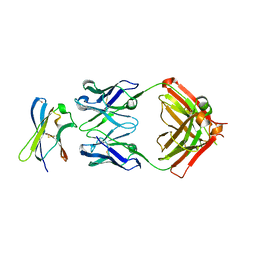 | | Crystal structure of Fab in complex with MICA alpha3 domain | | Descriptor: | 1,2-ETHANEDIOL, 2-acetamido-2-deoxy-beta-D-glucopyranose, Fab heavy chain, ... | | Authors: | Lee, P.S, Strop, P. | | Deposit date: | 2021-09-16 | | Release date: | 2022-10-05 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Residue-Level Characterization of Antibody Binding Epitopes Using Carbene Chemical Footprinting.
Anal.Chem., 95, 2023
|
|
4M5Y
 
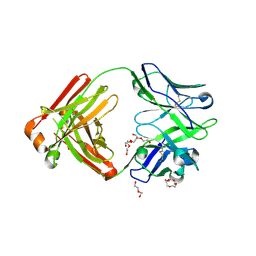 | | Crystal structure of broadly neutralizing Fab 5J8 | | Descriptor: | DI(HYDROXYETHYL)ETHER, Fab 5J8 heavy chain, Fab 5J8 light chain, ... | | Authors: | Lee, P.S, Wilson, I.A. | | Deposit date: | 2013-08-08 | | Release date: | 2013-09-25 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (1.55 Å) | | Cite: | Antibody Recognition of the Pandemic H1N1 Influenza Virus Hemagglutinin Receptor Binding Site.
J.Virol., 87, 2013
|
|
4NM8
 
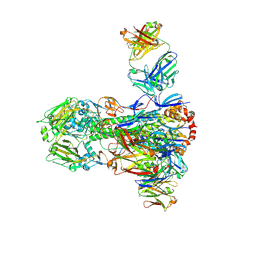 | |
4NM4
 
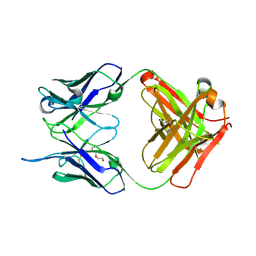 | |
5IC4
 
 | | Crystal structure of caspase-3 DEVE peptide complex | | Descriptor: | Caspase-3 subunit p12, Caspase-3 subunit p17, DEVE peptide | | Authors: | Seaman, J.E, Julien, O, Lee, P.S, Rettenmaier, T.J, Thomsen, N.D, Wells, J.A. | | Deposit date: | 2016-02-22 | | Release date: | 2016-07-20 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (2.65 Å) | | Cite: | Cacidases: caspases can cleave after aspartate, glutamate and phosphoserine residues.
Cell Death Differ., 23, 2016
|
|
5IC6
 
 | | Crystal structure of caspase-7 DEVE peptide complex | | Descriptor: | Caspase-7 subunit p11, Caspase-7 subunit p20, DEVE peptide | | Authors: | Seaman, J.E, Julien, O, Lee, P.S, Rettenmaier, T.J, Thomsen, N.D, Wells, J.A. | | Deposit date: | 2016-02-22 | | Release date: | 2016-07-20 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | Cacidases: caspases can cleave after aspartate, glutamate and phosphoserine residues.
Cell Death Differ., 23, 2016
|
|
8SZY
 
 | |
5WKZ
 
 | | VH1-69 germline antibody predicted from CR6261 | | Descriptor: | Immunoglobulin heavy variable 1-69D,IgG H chain, Lambda-chain (AA -20 to 215), SULFATE ION | | Authors: | Lang, S, Lee, P.S. | | Deposit date: | 2017-07-25 | | Release date: | 2018-08-01 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (1.85 Å) | | Cite: | Anti-idiotypic antibody K1-18 engages VH1-69 precursor and affinity-matured, anti-stem antibodies through mimicry of the HA stem
To Be Published
|
|
2O9L
 
 | | AMBER refined NMR Structure of the Sigma-54 RpoN Domain Bound to the-24 Promoter Element | | Descriptor: | 5'-D(*GP*AP*AP*AP*CP*GP*TP*GP*CP*CP*AP*AP*AP*A)-3', 5'-D(*TP*TP*TP*TP*GP*GP*CP*AP*CP*GP*TP*TP*TP*C)-3', RNA polymerase sigma factor RpoN | | Authors: | Doucleff, M, Pelton, J.G, Lee, P.S, Wemmer, D.E. | | Deposit date: | 2006-12-13 | | Release date: | 2007-07-17 | | Last modified: | 2023-12-27 | | Method: | SOLUTION NMR | | Cite: | Structural basis of DNA recognition by the alternative sigma-factor, sigma54.
J.Mol.Biol., 369, 2007
|
|
2O8K
 
 | | NMR Structure of the Sigma-54 RpoN Domain Bound to the-24 Promoter Element | | Descriptor: | 5'-D(*GP*AP*AP*AP*CP*GP*TP*GP*CP*CP*AP*AP*AP*A)-3', 5'-D(*TP*TP*TP*TP*GP*GP*CP*AP*CP*GP*TP*TP*TP*C)-3', RNA polymerase sigma factor RpoN | | Authors: | Doucleff, M, Pelton, J.G, Lee, P.S, Wemmer, D.E. | | Deposit date: | 2006-12-12 | | Release date: | 2007-07-17 | | Last modified: | 2023-12-27 | | Method: | SOLUTION NMR | | Cite: | Structural basis of DNA recognition by the alternative sigma-factor, sigma54.
J.Mol.Biol., 369, 2007
|
|
4M5Z
 
 | | Crystal structure of broadly neutralizing antibody 5J8 bound to 2009 pandemic influenza hemagglutinin, HA1 subunit | | Descriptor: | Fab 5J8 heavy chain, Fab 5J8 light chain, Hemagglutinin HA1 chain | | Authors: | Hong, M, Lee, P.S, Wilson, I.A. | | Deposit date: | 2013-08-08 | | Release date: | 2013-09-25 | | Last modified: | 2024-10-09 | | Method: | X-RAY DIFFRACTION (2.25 Å) | | Cite: | Antibody Recognition of the Pandemic H1N1 Influenza Virus Hemagglutinin Receptor Binding Site.
J.Virol., 87, 2013
|
|
