7L91
 
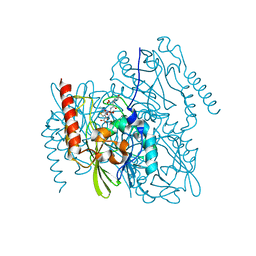 | | Structure of Metallo Beta-Lactamase L1 in a Complex with Hydrolyzed Moxalactam Determined by Pink-Beam Serial Crystallography | | Descriptor: | (2R)-2-[(R)-carboxy{[(2R)-2-carboxy-2-(4-hydroxyphenyl)acetyl]amino}methoxymethyl]-5-{[(1-methyl-1H-tetrazol-5-yl)sulfanyl]methyl}-3,6-dihydro-2H-1,3-oxazine-4-carboxylic acid, Putative metallo-beta-lactamase l1 (Beta-lactamase type ii) (Ec 3.5.2.6) (Penicillinase), ZINC ION | | Authors: | Wilamowski, M, Kim, Y, Sherrell, D.A, Lavens, A, Henning, R, Maltseva, N, Endres, M, Babnigg, G, Vukica, S, Joachimiak, A, Center for Structural Genomics of Infectious Diseases (CSGID) | | Deposit date: | 2021-01-01 | | Release date: | 2022-02-09 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Time-resolved beta-lactam cleavage by L1 metallo-beta-lactamase.
Nat Commun, 13, 2022
|
|
1VE6
 
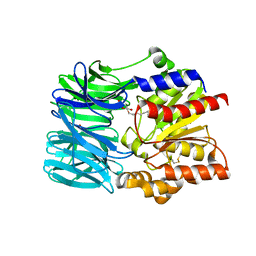 | | Crystal structure of an acylpeptide hydrolase/esterase from Aeropyrum pernix K1 | | Descriptor: | Acylamino-acid-releasing enzyme, GLYCEROL, octyl beta-D-glucopyranoside | | Authors: | Bartlam, M, Wang, G, Gao, R, Yang, H, Zhao, X, Xie, G, Cao, S, Feng, Y, Rao, Z. | | Deposit date: | 2004-03-27 | | Release date: | 2004-11-02 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Crystal structure of an acylpeptide hydrolase/esterase from Aeropyrum pernix K1
STRUCTURE, 12, 2004
|
|
6XIP
 
 | | The 1.5 A Crystal Structure of the Co-factor Complex of NSP7 and the C-terminal Domain of NSP8 from SARS CoV-2 | | Descriptor: | 1,2-ETHANEDIOL, Non-structural protein 7, Non-structural protein 8 | | Authors: | Wilamowski, M, Kim, Y, Jedrzejczak, R, Maltseva, N, Endres, M, Godzik, A, Michalska, K, Joachimiak, A, Center for Structural Genomics of Infectious Diseases (CSGID) | | Deposit date: | 2020-06-20 | | Release date: | 2020-07-01 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | Transient and stabilized complexes of Nsp7, Nsp8, and Nsp12 in SARS-CoV-2 replication.
Biophys.J., 120, 2021
|
|
6WIQ
 
 | | Crystal structure of the co-factor complex of NSP7 and the C-terminal domain of NSP8 from SARS CoV-2 | | Descriptor: | Non-structural protein 7, Non-structural protein 8 | | Authors: | Wilamowski, M, Kim, Y, Jedrzejczak, R, Maltseva, N, Endres, M, Godzik, A, Michalska, K, Joachimiak, A, Center for Structural Genomics of Infectious Diseases (CSGID) | | Deposit date: | 2020-04-10 | | Release date: | 2020-04-22 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (2.85 Å) | | Cite: | Transient and stabilized complexes of Nsp7, Nsp8, and Nsp12 in SARS-CoV-2 replication.
Biophys.J., 120, 2021
|
|
6XKM
 
 | | Room Temperature Structure of SARS-CoV-2 NSP10/NSP16 Methyltransferase in a Complex with SAM Determined by Fixed-Target Serial Crystallography | | Descriptor: | 2'-O-methyltransferase, CHLORIDE ION, Non-structural protein 10, ... | | Authors: | Wilamowski, M, Sherrell, D.A, Minasov, G, Kim, Y, Shuvalova, L, Lavens, A, Chard, R, Rosas-Lemus, M, Maltseva, N, Jedrzejczak, R, Michalska, K, Satchell, K.J.F, Joachimiak, A, Center for Structural Genomics of Infectious Diseases (CSGID) | | Deposit date: | 2020-06-26 | | Release date: | 2020-07-08 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (2.25 Å) | | Cite: | 2'-O methylation of RNA cap in SARS-CoV-2 captured by serial crystallography.
Proc.Natl.Acad.Sci.USA, 118, 2021
|
|
6WTC
 
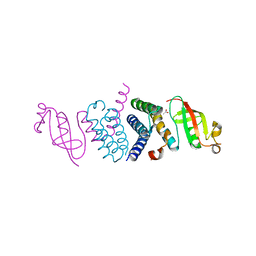 | | Crystal Structure of the Second Form of the Co-factor Complex of NSP7 and the C-terminal Domain of NSP8 from SARS CoV-2 | | Descriptor: | ACETIC ACID, Non-structural protein 7, Non-structural protein 8 | | Authors: | Wilamowski, M, Kim, Y, Jedrzejczak, R, Maltseva, N, Endres, M, Godzik, A, Michalska, K, Joachimiak, A, Center for Structural Genomics of Infectious Diseases (CSGID) | | Deposit date: | 2020-05-02 | | Release date: | 2020-05-13 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (1.85 Å) | | Cite: | Crystal Structure of the Second Form of the Co-factor Complex of NSP7 and the C-terminal Domain of NSP8 from SARS CoV-2
To Be Published
|
|
6SG4
 
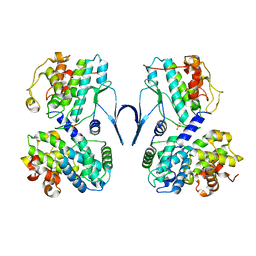 | | Structure of CDK2/cyclin A M246Q, S247EN | | Descriptor: | Cyclin-A2, Cyclin-dependent kinase 2 | | Authors: | Salamina, M, Basle, A, Massa, B, Noble, M.E.M, Endicott, J.A. | | Deposit date: | 2019-08-02 | | Release date: | 2021-01-27 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (2.43 Å) | | Cite: | Discriminative SKP2 Interactions with CDK-Cyclin Complexes Support a Cyclin A-Specific Role in p27KIP1 Degradation.
J.Mol.Biol., 433, 2021
|
|
1I2U
 
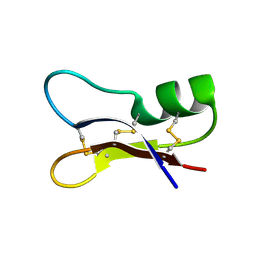 | | NMR SOLUTION STRUCTURES OF ANTIFUNGAL HELIOMICIN | | Descriptor: | DEFENSIN HELIOMICIN | | Authors: | Lamberty, M, Caille, A, Landon, C, Tassin-Moindrot, S, Hetru, C, Bulet, P, Vovelle, F. | | Deposit date: | 2001-02-12 | | Release date: | 2002-02-12 | | Last modified: | 2022-02-23 | | Method: | SOLUTION NMR | | Cite: | Solution structures of the antifungal heliomicin and a selected variant with both antibacterial and antifungal activities.
Biochemistry, 40, 2001
|
|
1NTL
 
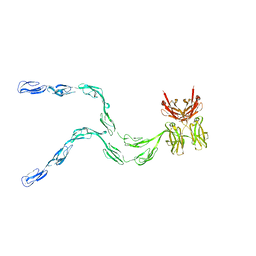 | | Model of mouse Crry-Ig determined by solution scattering, curve fitting and homology modelling | | Descriptor: | Complement component receptor 1-like protein,Ig gamma-1 chain C region secreted form | | Authors: | Aslam, M, Guthridge, J.M, Hack, B.K, Quigg, R.J, Holers, V.M, Perkins, S.J. | | Deposit date: | 2003-01-30 | | Release date: | 2004-02-03 | | Last modified: | 2024-02-14 | | Method: | SOLUTION SCATTERING (30 Å) | | Cite: | The extended multidomain solution structures of the complement protein Crry
and its chimaeric conjugate Crry-Ig by scattering, analytical ultracentrifugation
and constrained modelling: implications for function and therapy
J.Mol.Biol., 329, 2003
|
|
1NTJ
 
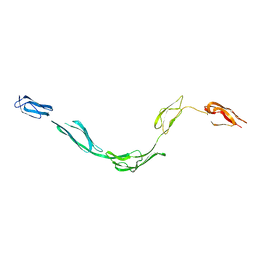 | | Model of rat Crry determined by solution scattering, curve fitting and homology modelling | | Descriptor: | complement receptor related protein | | Authors: | Aslam, M, Guthridge, J.M, Hack, B.K, Quigg, R.J, Holers, V.M, Perkins, S.J. | | Deposit date: | 2003-01-30 | | Release date: | 2004-02-03 | | Last modified: | 2024-02-14 | | Method: | SOLUTION SCATTERING (30 Å) | | Cite: | The Extended Multidomain Solution Structures of the Complement Protein
Crry and its Chimeric Conjugate Crry-Ig by Scattering, Analytical
Ultracentrifugation and Constrained Modelling: Implications for Function and
Therapy
J.Mol.Biol., 329, 2003
|
|
1HAQ
 
 | |
1I2V
 
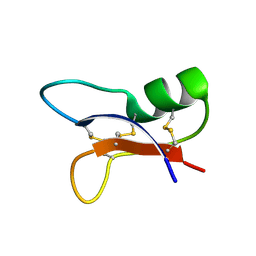 | | NMR SOLUTION STRUCTURES OF AN ANTIFUNGAL AND ANTIBACTERIAL MUTANT OF HELIOMICIN | | Descriptor: | DEFENSIN HELIOMICIN | | Authors: | Lamberty, M, Caille, A, Landon, C, Tassin-Moindrot, S, Hetru, C, Bulet, P, Vovelle, F. | | Deposit date: | 2001-02-12 | | Release date: | 2002-02-12 | | Last modified: | 2021-10-27 | | Method: | SOLUTION NMR | | Cite: | Solution structures of the antifungal heliomicin and a selected variant with both antibacterial and antifungal activities.
Biochemistry, 40, 2001
|
|
1VE7
 
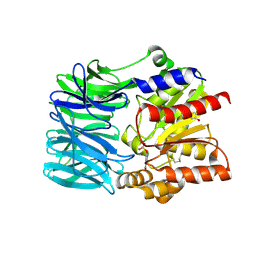 | | Crystal structure of an acylpeptide hydrolase/esterase from Aeropyrum pernix K1 in complex with p-nitrophenyl phosphate | | Descriptor: | 4-NITROPHENYL PHOSPHATE, Acylamino-acid-releasing enzyme, GLYCEROL | | Authors: | Bartlam, M, Wang, G, Gao, R, Yang, H, Zhao, X, Xie, G, Cao, S, Feng, Y, Rao, Z. | | Deposit date: | 2004-03-27 | | Release date: | 2004-11-02 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | Crystal structure of an acylpeptide hydrolase/esterase from Aeropyrum pernix K1
STRUCTURE, 12, 2004
|
|
2BAP
 
 | |
3EG5
 
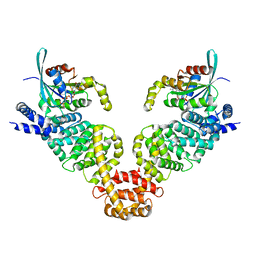 | | Crystal structure of MDIA1-TSH GBD-FH3 in complex with CDC42-GMPPNP | | Descriptor: | Cell division control protein 42 homolog, MAGNESIUM ION, PHOSPHOAMINOPHOSPHONIC ACID-GUANYLATE ESTER, ... | | Authors: | Lammers, M, Meyer, S, Kuehlmann, D, Wittinghofer, A. | | Deposit date: | 2008-09-10 | | Release date: | 2008-10-14 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | Specificity of Interactions between mDia Isoforms and Rho Proteins
J.Biol.Chem., 283, 2008
|
|
2X2A
 
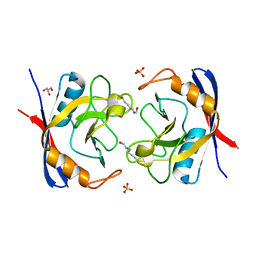 | | Free acetyl-CypA trigonal form | | Descriptor: | GLYCEROL, PEPTIDYL-PROLYL CIS-TRANS ISOMERASE A, SULFATE ION | | Authors: | Lammers, M, Neumann, H, Chin, J.W, James, L.C. | | Deposit date: | 2010-01-12 | | Release date: | 2010-03-23 | | Last modified: | 2011-07-13 | | Method: | X-RAY DIFFRACTION (1.4 Å) | | Cite: | Acetylation Regulates Cyclophilin a Catalysis, Immunosuppression and HIV Isomerization.
Nat.Chem.Biol., 6, 2010
|
|
2X2D
 
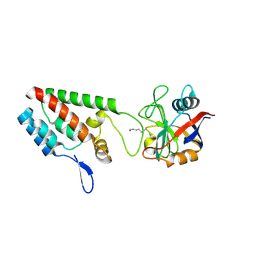 | | acetyl-CypA:HIV-1 N-term capsid domain complex | | Descriptor: | CAPSID PROTEIN P24, PEPTIDYL-PROLYL CIS-TRANS ISOMERASE A | | Authors: | Lammers, M, Neumann, H, Chin, J.W, James, L.C. | | Deposit date: | 2010-01-12 | | Release date: | 2010-03-23 | | Last modified: | 2018-01-24 | | Method: | X-RAY DIFFRACTION (1.95 Å) | | Cite: | Acetylation Regulates Cyclophilin a Catalysis, Immunosuppression and HIV Isomerisation
Nat.Chem.Biol., 6, 2010
|
|
2X2C
 
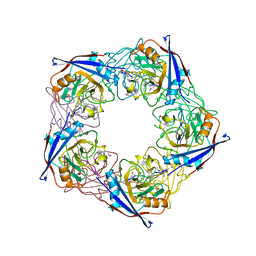 | | acetyl-CypA:cyclosporine complex | | Descriptor: | CYCLOSPORIN A, PEPTIDYL-PROLYL CIS-TRANS ISOMERASE A | | Authors: | Lammers, M, Neumann, H, Chin, J.W, James, L.C. | | Deposit date: | 2010-01-12 | | Release date: | 2010-03-23 | | Last modified: | 2018-05-02 | | Method: | X-RAY DIFFRACTION (2.41 Å) | | Cite: | Acetylation Regulates Cyclophilin a Catalysis, Immunosuppression and HIV Isomerisation
Nat.Chem.Biol., 6, 2010
|
|
2X25
 
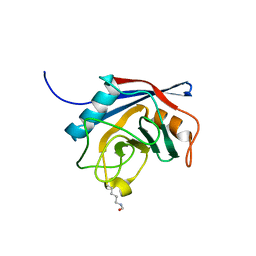 | | Free acetyl-CypA orthorhombic form | | Descriptor: | PEPTIDYL-PROLYL CIS-TRANS ISOMERASE A | | Authors: | Lammers, M, Neumann, H, Chin, J.W, James, L.C. | | Deposit date: | 2010-01-11 | | Release date: | 2010-03-23 | | Last modified: | 2011-07-13 | | Method: | X-RAY DIFFRACTION (1.2 Å) | | Cite: | Acetylation Regulates Cyclophilin a Catalysis, Immunosuppression and HIV Isomerization.
Nat.Chem.Biol., 6, 2010
|
|
7JIB
 
 | | Room Temperature Crystal Structure of Nsp10/Nsp16 from SARS-CoV-2 with Substrates and Products of 2'-O-methylation of the Cap-1 | | Descriptor: | 2'-O-methyltransferase, 7-METHYL-GUANOSINE-5'-TRIPHOSPHATE, 7-METHYL-GUANOSINE-5'-TRIPHOSPHATE-5'-(2'-O-METHYL)-ADENOSINE, ... | | Authors: | Wilamowski, M, Minasov, G, Kim, Y, Sherrell, D.A, Shuvalova, L, Lavens, A, Chard, R, Rosas-Lemus, M, Maltseva, N, Jedrzejczak, R, Michalska, K, Satchell, K.J.F, Joachimiak, A, Center for Structural Genomics of Infectious Diseases (CSGID) | | Deposit date: | 2020-07-23 | | Release date: | 2020-08-26 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (2.65 Å) | | Cite: | 2'-O methylation of RNA cap in SARS-CoV-2 captured by serial crystallography.
Proc.Natl.Acad.Sci.USA, 118, 2021
|
|
7KOA
 
 | | Room Temperature Structure of SARS-CoV-2 Nsp10/16 Methyltransferase in a Complex with Cap-0 and SAM Determined by Pink-Beam Serial Crystallography | | Descriptor: | 2'-O-methyltransferase, Non-structural protein 10, P1-7-METHYLGUANOSINE-P3-ADENOSINE-5',5'-TRIPHOSPHATE, ... | | Authors: | Wilamowski, M, Sherrell, D.A, Minasov, G, Shuvalova, L, Lavens, A, Henning, R, Maltseva, N, Rosas-Lemus, M, Kim, Y, Satchell, K.J.F, Srajer, V, Joachimiak, A, Center for Structural Genomics of Infectious Diseases (CSGID) | | Deposit date: | 2020-11-07 | | Release date: | 2020-11-18 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Room Temperature Structure of SARS-CoV-2 Nsp10/Nsp16 Methyltransferase in a Complex with Cap-0 and SAM Determined by Pink-Beam Serial Crystallography
To Be Published
|
|
7JPE
 
 | | Room Temperature Structure of SARS-CoV-2 Nsp10/Nsp16 Methyltransferase in a Complex with m7GpppA Cap-0 and SAM Determined by Fixed-Target Serial Crystallography | | Descriptor: | 2'-O-methyltransferase, 7N-METHYL-8-HYDROGUANOSINE-5'-DIPHOSPHATE, Non-structural protein 10, ... | | Authors: | Wilamowski, M, Sherrell, D.A, Minasov, G, Kim, Y, Shuvalova, L, Lavens, A, Chard, R, Rosas-Lemus, M, Maltseva, N, Jedrzejczak, R, Michalska, K, Satchell, K.J.F, Joachimiak, A, Center for Structural Genomics of Infectious Diseases (CSGID) | | Deposit date: | 2020-08-07 | | Release date: | 2020-08-26 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (2.18 Å) | | Cite: | 2'-O methylation of RNA cap in SARS-CoV-2 captured by serial crystallography.
Proc.Natl.Acad.Sci.USA, 118, 2021
|
|
7JHE
 
 | | Room Temperature Structure of SARS-CoV-2 Nsp10/Nsp16 Methyltransferase in a Complex with 2'-O-methylated m7GpppA Cap-1 and SAH Determined by Fixed-Target Serial Crystallography | | Descriptor: | 2'-O-methyltransferase, 7-METHYL-GUANOSINE-5'-TRIPHOSPHATE, 7-METHYL-GUANOSINE-5'-TRIPHOSPHATE-5'-(2'-O-METHYL)-ADENOSINE, ... | | Authors: | Wilamowski, M, Sherrell, D.A, Minasov, G, Kim, Y, Shuvalova, L, Lavens, A, Chard, R, Rosas-Lemus, M, Maltseva, N, Jedrzejczak, R, Michalska, K, Satchell, K.J.F, Joachimiak, A, Center for Structural Genomics of Infectious Diseases (CSGID) | | Deposit date: | 2020-07-20 | | Release date: | 2020-08-26 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (2.25 Å) | | Cite: | 2'-O methylation of RNA cap in SARS-CoV-2 captured by serial crystallography.
Proc.Natl.Acad.Sci.USA, 118, 2021
|
|
7L52
 
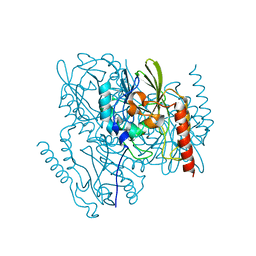 | | Crystal Structure of the Metallo Beta Lactamase L1 from Stenotrophomonas maltophilia Determined by Serial Crystallography | | Descriptor: | Putative metallo-beta-lactamase l1 (Beta-lactamase type ii) (Ec 3.5.2.6) (Penicillinase), ZINC ION | | Authors: | Wilamowski, M, Kim, Y, Sherrell, D.A, Lavens, A, Maltseva, N, Endres, M, Babnigg, G, Joachimiak, A, Center for Structural Genomics of Infectious Diseases (CSGID) | | Deposit date: | 2020-12-21 | | Release date: | 2020-12-30 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (1.85 Å) | | Cite: | Crystal Structure of the Metallo Beta Lactamase L1 from Stenotrophomonas maltophilia Determined by Serial Crystallography
To Be Published
|
|
7UHN
 
 | | Time-Resolved Structure of Metallo Beta-Lactamase L1 in a Complex with Cleaved Moxalactam (300 ms Snapshot) | | Descriptor: | (2R)-2-[(R)-carboxy{[(2R)-2-carboxy-2-(4-hydroxyphenyl)acetyl]amino}methoxymethyl]-5-{[(1-methyl-1H-tetrazol-5-yl)sulfanyl]methyl}-3,6-dihydro-2H-1,3-oxazine-4-carboxylic acid, Putative metallo-beta-lactamase l1 (Beta-lactamase type ii) (Ec 3.5.2.6) (Penicillinase), ZINC ION | | Authors: | Wilamowski, M, Kim, Y, Sherrell, D.A, Lavens, A, Henning, R, Maltseva, N, Endres, M, Babnigg, G, Srajer, V, Joachimiak, A, Center for Structural Genomics of Infectious Diseases (CSGID) | | Deposit date: | 2022-03-27 | | Release date: | 2022-04-13 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Time-resolved beta-lactam cleavage by L1 metallo-beta-lactamase.
Nat Commun, 13, 2022
|
|
