8DWI
 
 | |
4LRV
 
 | | Crystal structure of DndE from Escherichia coli B7A involved in DNA phosphorothioation modification | | Descriptor: | DNA sulfur modification protein DndE | | Authors: | Hu, W, Wang, C.K, Liang, J.D, Zhang, T.L, Yang, M, Hu, Z.P, Wang, Z.J, Lan, W.X, Wu, H.M, Ding, J.P, Wu, G, Deng, Z.X, Cao, C. | | Deposit date: | 2013-07-21 | | Release date: | 2013-08-28 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Structural insights into DndE from Escherichia coli B7A involved in DNA phosphorothioation modification
Cell Res., 22, 2012
|
|
6URT
 
 | |
8TNY
 
 | |
8TNW
 
 | |
8TNX
 
 | |
8VSI
 
 | | Mechanistic Insights Revealed by YbtPQ in the Occluded State | | Descriptor: | ABC transporter ATP-binding protein, ADP ORTHOVANADATE, MAGNESIUM ION, ... | | Authors: | Hu, W, Parkinson, C, Zheng, H. | | Deposit date: | 2024-01-24 | | Release date: | 2024-04-10 | | Method: | ELECTRON MICROSCOPY (3.1 Å) | | Cite: | Mechanistic Insights Revealed by YbtPQ in the Occluded State.
Biomolecules, 14, 2024
|
|
7LB8
 
 | | Structure of a ferrichrome importer FhuCDB from E. coli | | Descriptor: | Iron(3+)-hydroxamate import ATP-binding protein FhuC, Iron(3+)-hydroxamate import system permease protein FhuB, Iron(3+)-hydroxamate-binding protein FhuD | | Authors: | Hu, W, Zheng, H. | | Deposit date: | 2021-01-07 | | Release date: | 2021-11-24 | | Last modified: | 2024-05-29 | | Method: | ELECTRON MICROSCOPY (3.4 Å) | | Cite: | Cryo-EM reveals unique structural features of the FhuCDB Escherichia coli ferrichrome importer.
Commun Biol, 4, 2021
|
|
7CT1
 
 | |
8H0L
 
 | | Sulfur binding domain of Hga complexed with phosphorothioated DNA | | Descriptor: | DNA (5'-D(*CP*GP*AP*GP*(PST)P*TP*CP*GP*GP*C)-3'), DNA (5'-D(*GP*CP*CP*GP*AP*AP*CP*TP*CP*G)-3'), MAGNESIUM ION, ... | | Authors: | Liu, G, He, X, Hu, W, Yang, B, Xiao, Q. | | Deposit date: | 2022-09-29 | | Release date: | 2023-09-27 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Characterization of a promiscuous DNA sulfur binding domain and application in site-directed RNA base editing.
Nucleic Acids Res., 51, 2023
|
|
1EXY
 
 | | SOLUTION STRUCTURE OF HTLV-1 PEPTIDE BOUND TO ITS RNA APTAMER TARGET | | Descriptor: | HTLV-1 REX PEPTIDE, RNA APTAMER, 33-MER | | Authors: | Jiang, F, Gorin, A, Hu, W, Majumdar, A, Baskerville, S, Xu, W, Ellington, A, Patel, D.J. | | Deposit date: | 2000-05-05 | | Release date: | 2000-05-15 | | Last modified: | 2024-05-22 | | Method: | SOLUTION NMR | | Cite: | Anchoring an extended HTLV-1 Rex peptide within an RNA major groove containing junctional base triples.
Structure Fold.Des., 7, 1999
|
|
2BPR
 
 | | NMR STRUCTURE OF THE SUBSTRATE BINDING DOMAIN OF DNAK, 25 STRUCTURES | | Descriptor: | DNAK | | Authors: | Wang, H, Kurochkin, A.V, Pang, Y, Hu, W, Flynn, G.C, Zuiderweg, E.R.P. | | Deposit date: | 1998-08-11 | | Release date: | 1999-03-02 | | Last modified: | 2024-05-22 | | Method: | SOLUTION NMR | | Cite: | NMR solution structure of the 21 kDa chaperone protein DnaK substrate binding domain: a preview of chaperone-protein interaction.
Biochemistry, 37, 1998
|
|
6P6I
 
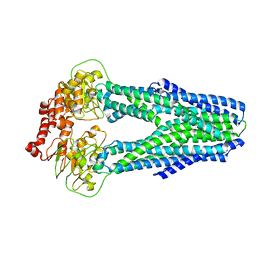 | | Structure of YbtPQ importer | | Descriptor: | ABC transporter protein, inner membrane ABC-transporter | | Authors: | Wang, Z, Hu, W, Zheng, H. | | Deposit date: | 2019-06-04 | | Release date: | 2020-03-04 | | Last modified: | 2024-03-13 | | Method: | ELECTRON MICROSCOPY (3.67 Å) | | Cite: | Pathogenic siderophore ABC importer YbtPQ adopts a surprising fold of exporter.
Sci Adv, 6, 2020
|
|
9NM2
 
 | | Dimeric Structure of full-length CrgA, a Cell Division Protein from Mycobacterium tuberculosis, in Lipid Bilayers | | Descriptor: | Cell division protein CrgA | | Authors: | Shin, Y, Prasad, R, Das, N, Taylor, J.A, Qin, H, Hu, W, Hu, Y.-Y, Fu, R, Zhang, R, Zhou, H.-X, Cross, T.A. | | Deposit date: | 2025-03-03 | | Release date: | 2025-04-02 | | Method: | SOLID-STATE NMR | | Cite: | Mycobacterium tuberculosis CrgA Forms a Dimeric Structure with Its Transmembrane Domain Sandwiched between Cytoplasmic and Periplasmic beta-Sheets, Enabling Multiple Interactions with Other Divisome Proteins.
J.Am.Chem.Soc., 2025
|
|
1G70
 
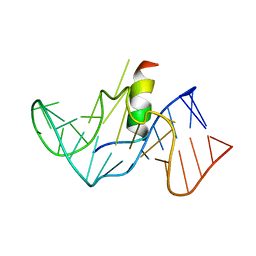 | | COMPLEX OF HIV-1 RRE-IIB RNA WITH RSG-1.2 PEPTIDE | | Descriptor: | HIV-1 RRE-IIB 32 NUCLEOTIDE RNA, RSG-1.2 PEPTIDE | | Authors: | Gosser, Y, Hermann, T, Majumdar, A, Hu, W, Frederick, R, Jiang, F, Xu, W, Patel, D.J. | | Deposit date: | 2000-11-08 | | Release date: | 2001-02-07 | | Last modified: | 2024-05-22 | | Method: | SOLUTION NMR | | Cite: | Peptide-triggered conformational switch in HIV-1 RRE RNA complexes.
Nat.Struct.Biol., 8, 2001
|
|
1NEM
 
 | | Saccharide-RNA recognition in the neomycin B / RNA aptamer complex | | Descriptor: | 2,6-diamino-2,6-dideoxy-alpha-D-glucopyranose, 2,6-diamino-2,6-dideoxy-beta-L-idopyranose-(1-3)-beta-D-ribofuranose, 2-DEOXY-D-STREPTAMINE, ... | | Authors: | Jiang, L, Majumdar, A, Hu, W, Jaishree, T.J, Xu, W, Patel, D.J. | | Deposit date: | 1999-03-15 | | Release date: | 1999-08-31 | | Last modified: | 2023-12-27 | | Method: | SOLUTION NMR | | Cite: | Saccharide-RNA recognition in a complex formed between neomycin B and an RNA aptamer
Structure Fold.Des., 7, 1999
|
|
1UMT
 
 | | Stromelysin-1 catalytic domain with hydrophobic inhibitor bound, ph 7.0, 32oc, 20 mm cacl2, 15% acetonitrile; nmr average of 20 structures minimized with restraints | | Descriptor: | CALCIUM ION, N-{(2R)-2-[2-(hydroxyamino)-2-oxoethyl]-4-methylpentanoyl}-L-leucyl-L-phenylalaninamide, STROMELYSIN-1, ... | | Authors: | Van Doren, S.R, Kurochkin, A.V, Hu, W, Zuiderweg, E.R.P. | | Deposit date: | 1995-10-31 | | Release date: | 1996-03-08 | | Last modified: | 2024-05-01 | | Method: | SOLUTION NMR | | Cite: | Solution structure of the catalytic domain of human stromelysin complexed with a hydrophobic inhibitor.
Protein Sci., 4, 1995
|
|
1UMS
 
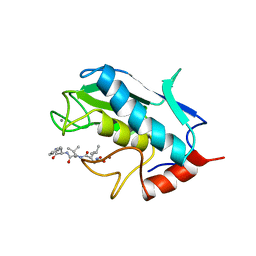 | | STROMELYSIN-1 CATALYTIC DOMAIN WITH HYDROPHOBIC INHIBITOR BOUND, PH 7.0, 32OC, 20 MM CACL2, 15% ACETONITRILE; NMR ENSEMBLE OF 20 STRUCTURES | | Descriptor: | CALCIUM ION, N-{(2R)-2-[2-(hydroxyamino)-2-oxoethyl]-4-methylpentanoyl}-L-leucyl-L-phenylalaninamide, STROMELYSIN-1, ... | | Authors: | Van Doren, S.R, Kurochkin, A.V, Hu, W, Zuiderweg, E.R.P. | | Deposit date: | 1995-10-31 | | Release date: | 1996-03-08 | | Last modified: | 2024-05-01 | | Method: | SOLUTION NMR | | Cite: | Solution structure of the catalytic domain of human stromelysin complexed with a hydrophobic inhibitor.
Protein Sci., 4, 1995
|
|
1RW2
 
 | | Three-dimensional structure of Ku80 CTD | | Descriptor: | ATP-dependent DNA helicase II, 80 kDa subunit | | Authors: | Zhang, Z, Hu, W, Cano, L, Lee, T.D, Chen, D.J, Chen, Y. | | Deposit date: | 2003-12-15 | | Release date: | 2003-12-30 | | Last modified: | 2024-05-22 | | Method: | SOLUTION NMR | | Cite: | Solution structure of the C-terminal domain of Ku80 suggests important sites for protein-protein interactions.
STRUCTURE, 12, 2004
|
|
484D
 
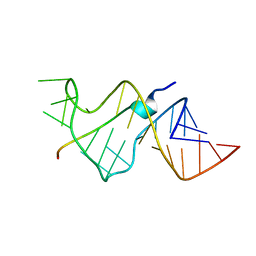 | | SOLUTION STRUCTURE OF HIV-1 REV PEPTIDE-RNA APTAMER COMPLEX | | Descriptor: | BASIC REV PEPTIDE, RNA APTAMER | | Authors: | Ye, X, Gorin, A.A, Frederick, R, Hu, W, Majumdar, A, Xu, W, Mclendon, G, Ellington, A, Patel, D.J. | | Deposit date: | 1999-08-02 | | Release date: | 1999-10-14 | | Last modified: | 2023-12-27 | | Method: | SOLUTION NMR | | Cite: | RNA architecture dictates the conformations of a bound peptide.
Chem.Biol., 6, 1999
|
|
1L4S
 
 | |
2LGG
 
 | | Structure of PHD domain of UHRF1 in complex with H3 peptide | | Descriptor: | E3 ubiquitin-protein ligase UHRF1, ZINC ION, histone H3 peptide | | Authors: | Wang, C, Shen, J, Yang, Z, Chen, P, Zhao, B, Hu, W, Lan, W, Tong, X, Wu, H, Li, G, Cao, C. | | Deposit date: | 2011-07-26 | | Release date: | 2011-09-28 | | Last modified: | 2024-05-15 | | Method: | SOLUTION NMR | | Cite: | Structural basis for site-specific reading of unmodified R2 of histone H3 tail by UHRF1 PHD finger.
Cell Res., 21, 2011
|
|
2LGL
 
 | | NMR structure of the UHRF1 PHD domain | | Descriptor: | E3 ubiquitin-protein ligase UHRF1, ZINC ION | | Authors: | Wang, C, Shen, J, Yang, Z, Chen, P, Zhao, B, Hu, W, Lan, W, Tong, X, Wu, H, Li, G, Cao, C. | | Deposit date: | 2011-07-28 | | Release date: | 2011-09-28 | | Last modified: | 2024-05-15 | | Method: | SOLUTION NMR | | Cite: | Structural basis for site-specific reading of unmodified R2 of histone H3 tail by UHRF1 PHD finger.
Cell Res., 21, 2011
|
|
2LGK
 
 | | NMR Structure of UHRF1 PHD domains in a complex with histone H3 peptide | | Descriptor: | E3 ubiquitin-protein ligase UHRF1, ZINC ION, histone H3 peptide | | Authors: | Wang, C, Shen, J, Yang, Z, Chen, P, Zhao, B, Hu, W, Lan, W, Tong, X, Wu, H, Li, G, Cao, C. | | Deposit date: | 2011-07-28 | | Release date: | 2011-09-28 | | Last modified: | 2023-06-14 | | Method: | SOLUTION NMR | | Cite: | Structural basis for site-specific reading of unmodified R2 of histone H3 tail by UHRF1 PHD finger.
Cell Res., 21, 2011
|
|
6P6J
 
 | | Structure of YbtPQ importer with substrate Ybt-Fe bound | | Descriptor: | ABC transporter protein, FE (III) ION, inner membrane ABC-transporter, ... | | Authors: | Wang, Z, Hu, W, Zheng, H. | | Deposit date: | 2019-06-04 | | Release date: | 2020-03-04 | | Last modified: | 2024-03-13 | | Method: | ELECTRON MICROSCOPY (3.4 Å) | | Cite: | Pathogenic siderophore ABC importer YbtPQ adopts a surprising fold of exporter.
Sci Adv, 6, 2020
|
|
