2Q9S
 
 | |
2QM9
 
 | | Troglitazone Bound to Fatty Acid Binding Protein 4 | | Descriptor: | (5R)-5-(4-{[(2R)-6-HYDROXY-2,5,7,8-TETRAMETHYL-3,4-DIHYDRO-2H-CHROMEN-2-YL]METHOXY}BENZYL)-1,3-THIAZOLIDINE-2,4-DIONE, Fatty acid-binding protein, adipocyte, ... | | Authors: | Gillilan, R.E, Ayers, S.D, Noy, N. | | Deposit date: | 2007-07-14 | | Release date: | 2007-10-09 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (2.31 Å) | | Cite: | Structural Basis for Activation of Fatty Acid-binding Protein 4
J.Mol.Biol., 372, 2007
|
|
3JBH
 
 | | TWO HEAVY MEROMYOSIN INTERACTING-HEADS MOTIFS FLEXIBLE DOCKED INTO TARANTULA THICK FILAMENT 3D-MAP ALLOWS IN DEPTH STUDY OF INTRA- AND INTERMOLECULAR INTERACTIONS | | Descriptor: | MYOSIN 2 ESSENTIAL LIGHT CHAIN STRIATED MUSCLE, MYOSIN 2 HEAVY CHAIN STRIATED MUSCLE, MYOSIN 2 REGULATORY LIGHT CHAIN STRIATED MUSCLE | | Authors: | Alamo, L, Qi, D, Wriggers, W, Pinto, A, Zhu, J, Bilbao, A, Gillilan, R.E, Hu, S, Padron, R. | | Deposit date: | 2015-09-01 | | Release date: | 2016-03-09 | | Last modified: | 2024-02-21 | | Method: | ELECTRON MICROSCOPY (20 Å) | | Cite: | Conserved Intramolecular Interactions Maintain Myosin Interacting-Heads Motifs Explaining Tarantula Muscle Super-Relaxed State Structural Basis.
J. Mol. Biol., 428, 2016
|
|
5TBY
 
 | | HUMAN BETA CARDIAC HEAVY MEROMYOSIN INTERACTING-HEADS MOTIF OBTAINED BY HOMOLOGY MODELING (USING SWISS-MODEL) OF HUMAN SEQUENCE FROM APHONOPELMA HOMOLOGY MODEL (PDB-3JBH), RIGIDLY FITTED TO HUMAN BETA-CARDIAC NEGATIVELY STAINED THICK FILAMENT 3D-RECONSTRUCTION (EMD-2240) | | Descriptor: | Myosin light chain 3, Myosin regulatory light chain 2, ventricular/cardiac muscle isoform, ... | | Authors: | ALAMO, L, WARE, J.S, PINTO, A, GILLILAN, R.E, SEIDMAN, J.G, SEIDMAN, C.E, PADRON, R. | | Deposit date: | 2016-09-13 | | Release date: | 2017-06-07 | | Last modified: | 2024-10-23 | | Method: | ELECTRON MICROSCOPY (20 Å) | | Cite: | Effects of myosin variants on interacting-heads motif explain distinct hypertrophic and dilated cardiomyopathy phenotypes.
Elife, 6, 2017
|
|
7UXE
 
 | | Pseudomonas phage E217 small terminase (TerS) | | Descriptor: | Small terminase | | Authors: | Lokareddy, R.K, Hou, C.-F.D, Doll, S.G, Li, F, Gillilan, R, Forti, F, Briani, F, Cingolani, G. | | Deposit date: | 2022-05-05 | | Release date: | 2022-09-28 | | Last modified: | 2024-06-12 | | Method: | ELECTRON MICROSCOPY (3.38 Å) | | Cite: | Terminase Subunits from the Pseudomonas-Phage E217.
J.Mol.Biol., 434, 2022
|
|
6W7T
 
 | | Structure of PaP3 small terminase | | Descriptor: | small terminase subunit | | Authors: | Cingolani, G, Lokareddy, R. | | Deposit date: | 2020-03-19 | | Release date: | 2020-11-11 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (3.01 Å) | | Cite: | Biophysical analysis of Pseudomonas-phage PaP3 small terminase suggests a mechanism for sequence-specific DNA-binding by lateral interdigitation.
Nucleic Acids Res., 48, 2020
|
|
8DKR
 
 | |
7BDN
 
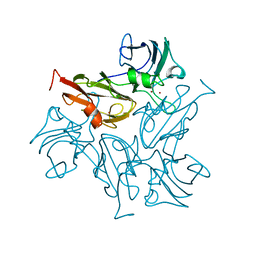 | | Structure of the Streptomyces coelicolor small laccase - cubic crystal form | | Descriptor: | COPPER (II) ION, Putative copper oxidase | | Authors: | Zovo, K, Majumdar, S, Lukk, T. | | Deposit date: | 2020-12-22 | | Release date: | 2022-01-12 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | Substitution of the Methionine Axial Ligand of the T1 Copper for the Fungal-like Phenylalanine Ligand (M298F) Causes Local Structural Perturbations that Lead to Thermal Instability and Reduced Catalytic Efficiency of the Small Laccase from Streptomyces coelicolor A3(2).
Acs Omega, 7, 2022
|
|
7BFM
 
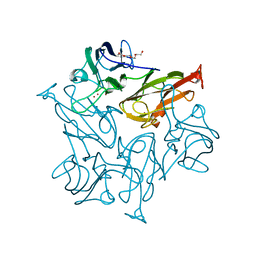 | |
6E3B
 
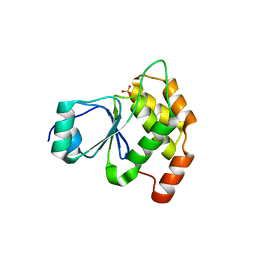 | |
7B2K
 
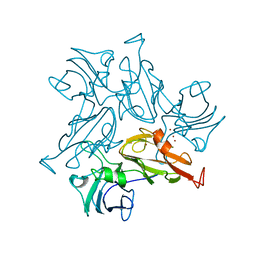 | |
7B4Y
 
 | |
7JOQ
 
 | | Structure of NV1 small terminase | | Descriptor: | Small Terminase subunit | | Authors: | Cingolani, G, Lokareddy, R. | | Deposit date: | 2020-08-07 | | Release date: | 2020-11-11 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (3.95 Å) | | Cite: | Biophysical analysis of Pseudomonas-phage PaP3 small terminase suggests a mechanism for sequence-specific DNA-binding by lateral interdigitation.
Nucleic Acids Res., 48, 2020
|
|
7LF4
 
 | |
7LFC
 
 | |
7LEU
 
 | |
7LET
 
 | |
7LEQ
 
 | |
7N9H
 
 | |
5T94
 
 | | Crystal structure of Kap60 bound to yeast RCC1 (Prp20) | | Descriptor: | Guanine nucleotide exchange factor SRM1, Importin subunit alpha | | Authors: | Sankhala, R.S, Lokareddy, R.K, Pumroy, R.A, Cingolani, G. | | Deposit date: | 2016-09-09 | | Release date: | 2017-09-13 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (2.631 Å) | | Cite: | Three-dimensional context rather than NLS amino acid sequence determines importin alpha subtype specificity for RCC1.
Nat Commun, 8, 2017
|
|
5TBK
 
 | | Crystal structure of human importin a3 bound to RCC1 | | Descriptor: | Importin subunit alpha-3, Regulator of chromosome condensation | | Authors: | Sankhala, R.S, Lokareddy, R.K, Pumroy, R.A, Cingolani, G. | | Deposit date: | 2016-09-12 | | Release date: | 2017-09-13 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (3.45 Å) | | Cite: | Three-dimensional context rather than NLS amino acid sequence determines importin alpha subtype specificity for RCC1.
Nat Commun, 8, 2017
|
|
