1ZPC
 
 | | Crystal Structure of the Catalytic Domain of Coagulation Factor XI in Complex with 2-[2-(3-Chloro-phenyl)-2-hydroxy-acetylamino]-N-[4-guanidino-1-(thiazole-2-carbonyl)-butyl]-3-methyl-butyramide | | Descriptor: | 2-[2-(3-CHLORO-PHENYL)-2-HYDROXY-ACETYLAMINO]-N-[4-GUANIDINO-1-(THIAZOLE-2-CARBONYL)-BUTYL]-3-METHYL-BUTYRAMIDE, Coagulation factor XI, SULFATE ION | | Authors: | Deng, H, Bannister, T.D, Jin, L, Nagafuji, P, Celatka, C.A, Lin, J, Lazarova, T.I, Rynkiewicz, M.J, Quinn, J, Bibbins, F, Pandey, P, Gorga, J, Babine, R.E, Meyers, H.V, Abdel-Meguid, S.S, Strickler, J.E. | | Deposit date: | 2005-05-16 | | Release date: | 2006-04-18 | | Last modified: | 2024-11-20 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Synthesis, SAR exploration, and X-ray crystal structures of factor XIa inhibitors containing an alpha-ketothiazole arginine
Bioorg.Med.Chem.Lett., 16, 2006
|
|
1ZPB
 
 | | Crystal Structure of the Catalytic Domain of Coagulation Factor XI in Complex with 4-Methyl-pentanoic acid {1-[4-guanidino-1-(thiazole-2-carbonyl)-butylcarbamoyl]-2-methyl-propyl}-amide | | Descriptor: | 4-METHYL-PENTANOIC ACID {1-[4-GUANIDINO-1-(THIAZOLE-2-CARBONYL)-BUTYLCARBAMOYL]-2-METHYL-PROPYL}-AMIDE, Coagulation factor XI, SULFATE ION | | Authors: | Deng, H, Bannister, T.D, Jin, L, Nagafuji, P, Celatka, C.A, Lin, J, Lazarova, T.I, Rynkiewicz, M.J, Quinn, J, Bibbins, F, Pandey, P, Gorga, J, Babine, R.E, Meyers, H.V, Abdel-Meguid, S.S, Strickler, J.E. | | Deposit date: | 2005-05-16 | | Release date: | 2006-04-18 | | Last modified: | 2024-11-20 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Synthesis, SAR exploration, and X-ray crystal structures of factor XIa inhibitors containing an alpha-ketothiazole arginine
Bioorg.Med.Chem.Lett., 16, 2006
|
|
2WR8
 
 | | Structure of Pyrococcus horikoshii SAM hydroxide adenosyltransferase in complex with SAH | | Descriptor: | PUTATIVE UNCHARACTERIZED PROTEIN PH0463, S-ADENOSYL-L-HOMOCYSTEINE | | Authors: | McMahon, S.A, Deng, H, O'Hagan, D, Johnson, K.A, Naismith, J.H. | | Deposit date: | 2009-08-31 | | Release date: | 2009-09-22 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (1.77 Å) | | Cite: | Mechanistic Insights Into Water Activation in Sam Hydroxide Adenosyltransferase (Duf-62).
Chembiochem, 10, 2009
|
|
8PE9
 
 | | Complex between DDR1 DS-like domain and PRTH-101 Fab | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-[alpha-L-fucopyranose-(1-6)]2-acetamido-2-deoxy-beta-D-glucopyranose, CALCIUM ION, ... | | Authors: | Liu, J, Chiang, H, Xiong, W, Laurent, V, Griffiths, S.C, Duelfer, J, Deng, H, Sun, X, Yin, Y.W, Li, W, Audoly, L.P, An, Z, Schuerpf, T, Li, R, Zhang, N. | | Deposit date: | 2023-06-13 | | Release date: | 2023-06-28 | | Last modified: | 2024-11-20 | | Method: | X-RAY DIFFRACTION (3.152 Å) | | Cite: | A highly selective humanized DDR1 mAb reverses immune exclusion by disrupting collagen fiber alignment in breast cancer.
J Immunother Cancer, 11, 2023
|
|
8AQF
 
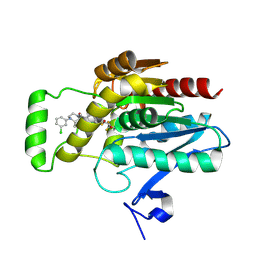 | | CRYSTAL STRUCTURE OF HUMAN MONOGLYCERIDE LIPASE WITH COMPOUND LEI-515 | | Descriptor: | 1-[(~{R})-[2-chloranyl-4-[(2~{S},3~{S})-4-(3-chlorophenyl)-2,3-dimethyl-piperazin-1-yl]carbonyl-phenyl]sulfinyl]-3,3-bis(fluoranyl)pentan-2-one, Monoglyceride lipase | | Authors: | Jiang, M, Huizenga, M, Wirt, J, Paloczi, J, Amedi, A, van der Berg, R, Benz, J, Collin, L, Deng, H, Driever, W, Florea, B, Grether, U, Janssen, A, Heitman, L, Lam, T.W, Mohr, F, Pavlovic, A, Ruf, I, Rutjes, H, Stevens, F, van der Vliet, D, van der Wel, T, Wittwer, M, Boeckel, C, Pacher, P, Hohmann, A, van der Stelt, M. | | Deposit date: | 2022-08-12 | | Release date: | 2023-08-23 | | Last modified: | 2025-03-12 | | Method: | X-RAY DIFFRACTION (1.55 Å) | | Cite: | A monoacylglycerol lipase inhibitor showing therapeutic efficacy in mice without central side effects or dependence.
Nat Commun, 14, 2023
|
|
1RQP
 
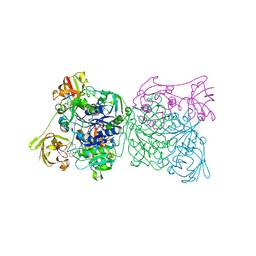 | | Crystal structure and mechanism of a bacterial fluorinating enzyme | | Descriptor: | 5'-fluoro-5'-deoxyadenosine synthase, S-ADENOSYLMETHIONINE | | Authors: | Dong, C, Huang, F, Deng, H, Schaffrath, C, Spencer, J.B, O'Hagan, D, Naismith, J.H. | | Deposit date: | 2003-12-06 | | Release date: | 2004-03-02 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Crystal structure and mechanism of a bacterial fluorinating enzyme
Nature, 427, 2004
|
|
1RQR
 
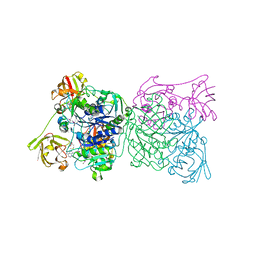 | | Crystal structure and mechanism of a bacterial fluorinating enzyme, product complex | | Descriptor: | 5'-FLUORO-5'-DEOXYADENOSINE, 5'-fluoro-5'-deoxyadenosine synthase, METHIONINE | | Authors: | Dong, C, Huang, F, Deng, H, Schaffrath, C, Spencer, J.B, O'Hagan, D, Naismith, J.H. | | Deposit date: | 2003-12-07 | | Release date: | 2004-03-02 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (2.67 Å) | | Cite: | Crystal structure and mechanism of a bacterial fluorinating enzyme
Nature, 427, 2004
|
|
1ZOM
 
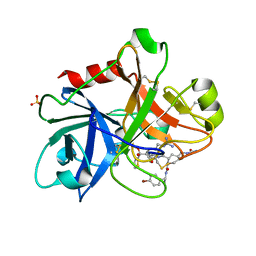 | | Crystal Structure of the Catalytic Domain of Coagulation Factor XI in complex with a peptidomimetic Inhibitor | | Descriptor: | (S)-2-(3-((R)-1-(4-BROMOPHENYL)ETHYL)UREIDO)-N-((S)-1-((S)-5-GUANIDINO-1-OXO-1-(THIAZOL-2-YL)PENTAN-2-YLAMINO)-3-METHYL-1-OXOBUTAN-2-YL)-5-UREIDOPENTANAMIDE, Coagulation factor XI, SULFATE ION | | Authors: | Lin, J, Deng, H, Jin, L, Pandey, P, Rynkiewicz, M, Bibbins, F, Cantin, S, Quinn, J, Magee, S, Gorga, J. | | Deposit date: | 2005-05-13 | | Release date: | 2006-05-23 | | Last modified: | 2024-11-20 | | Method: | X-RAY DIFFRACTION (2.25 Å) | | Cite: | Design, synthesis, and biological evaluation of peptidomimetic inhibitors of factor XIa as novel anticoagulants.
J.Med.Chem., 49, 2006
|
|
1DRO
 
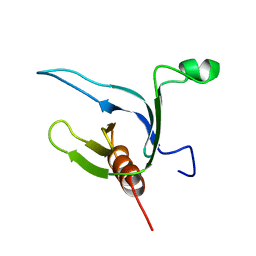 | | NMR STRUCTURE OF THE CYTOSKELETON/SIGNAL TRANSDUCTION PROTEIN | | Descriptor: | BETA-SPECTRIN | | Authors: | Zhang, P, Talluri, S, Deng, H, Branton, D, Wagner, G. | | Deposit date: | 1995-09-29 | | Release date: | 1996-04-03 | | Last modified: | 2024-05-22 | | Method: | SOLUTION NMR | | Cite: | Solution structure of the pleckstrin homology domain of Drosophila beta-spectrin.
Structure, 3, 1995
|
|
2C5H
 
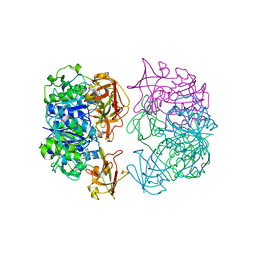 | | X-ray crystal structure of 5'-fluorodeoxyadenosine synthase from Streptomyces cattleya complexed with 2'deoxy-adenosine | | Descriptor: | (2R,3S,5R)-5-(6-amino-9H-purin-9-yl)-tetrahydro-2-(hydroxymethyl)furan-3-ol, 5'-FLUORO-5'-DEOXYADENOSINE SYNTHASE, CHLORIDE ION, ... | | Authors: | McEwan, A.R, Deng, H, McGlinchey, R.P, Robinson, D.R, O'Hagan, D, Naismith, J.H, Spencer, J. | | Deposit date: | 2005-10-27 | | Release date: | 2006-10-25 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | Substrates and Inhibitors of the Fluorinase from Streptomyces Cattleya
To be Published
|
|
2C4T
 
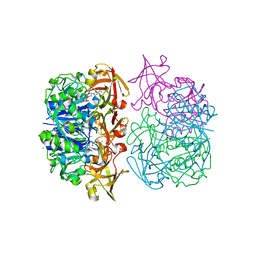 | | X-ray crystal structure of 5'-fluorodeoxyadenosine synthase from Streptomyces cattleya complexed with an inhibitor, an analogue of S- adenosyl methionine | | Descriptor: | 5'-FLUORO-5'-DEOXYADENOSINE SYNTHASE, CHLORIDE ION, S-5'-AZAMETHIONINE-5'-DEOXYADENOSINE | | Authors: | McEwan, A.R, Deng, H, McGlinchey, R.P, Robinson, D.R, O'Hagan, D, Naismith, J.H. | | Deposit date: | 2005-10-22 | | Release date: | 2007-03-27 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Substrates and Inhibitors of the Fluorinase from Streptomyces Cattleya
To be Published
|
|
2C4U
 
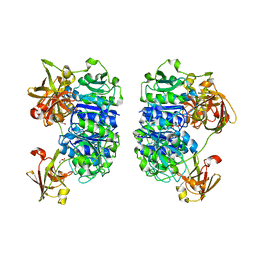 | | Crystal structure of the apo form of the 5'-Fluoro-5'-deoxyadenosine synthase enzyme from Streptomyces cattleya | | Descriptor: | 5'-FLUORO-5'-DEOXYADENOSINE SYNTHASE, GLYCEROL | | Authors: | McEwan, A.R, Deng, H, Robinson, D.A, DeLaurentis, W, McGlinchey, R.P, O'Hagan, D, Naismith, J.H. | | Deposit date: | 2005-10-22 | | Release date: | 2006-04-12 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Substrate specificity in enzymatic fluorination. The fluorinase from Streptomyces cattleya accepts 2'-deoxyadenosine substrates.
Org. Biomol. Chem., 4, 2006
|
|
2CC2
 
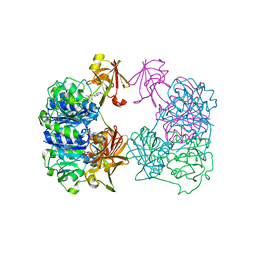 | | X-ray crystal structure of 5'-fluorodeoxyadenosine synthase from Streptomyces cattleya complexed with 5'deoxyadenosine | | Descriptor: | 5'-DEOXYADENOSINE, 5'-FLUORO-5'-DEOXYADENOSINE SYNTHASE, CHLORIDE ION | | Authors: | Mcewan, A.R, Deng, H, McGlinchey, R.P, Robinson, D.R, O'Hagan, D, Spencer, J, Naismith, J.H. | | Deposit date: | 2006-01-11 | | Release date: | 2007-03-20 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Substrate specificity in enzymatic fluorination. The fluorinase from Streptomyces cattleya accepts 2'-deoxyadenosine substrates.
Org. Biomol. Chem., 4, 2006
|
|
2C5B
 
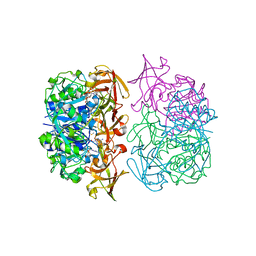 | | X-ray crystal structure of 5'-fluorodeoxyadenosine synthase from Streptomyces cattleya complexed with 2'deoxy-5'deoxy-fluoroadenosine. | | Descriptor: | 5'-FLUORO-2',5'-DIDEOXYADENOSINE, 5'-FLUORO-5'-DEOXYADENOSINE SYNTHASE, METHIONINE | | Authors: | McEwan, A.R, Deng, H, McGlinchey, R.P, Robinson, D.A, O'Hagan, D, Naismith, J.H, Spencer, J. | | Deposit date: | 2005-10-26 | | Release date: | 2007-02-20 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Substrate Specificity in Enzymatic Fluorination. The Fluorinase from Streptomyces Cattleya Accepts 2'-Deoxyadenosine Substrates.
Org.Biomol.Chem., 4, 2006
|
|
2CBX
 
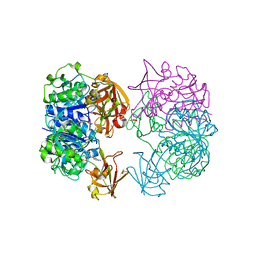 | | X-ray crystal structure of 5'-fluorodeoxyadenosine synthase from Streptomyces cattleya complexed with beta-D-erythrofuranosyl- adenosine | | Descriptor: | 5'-FLUORO-5'-DEOXYADENOSINE SYNTHASE, BETA-D-ERYTHROFURANOSYL-ADENOSINE, GLYCEROL | | Authors: | McEwan, A.R, Cadicamo, C.D, Deng, H, McGlinchey, R.P, Robinson, D.R, O'Hagan, D, Naismith, J.H, Spencer, J. | | Deposit date: | 2006-01-09 | | Release date: | 2007-03-06 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Substrate specificity in enzymatic fluorination. The fluorinase from Streptomyces cattleya accepts 2'-deoxyadenosine substrates.
Org. Biomol. Chem., 4, 2006
|
|
1ZPZ
 
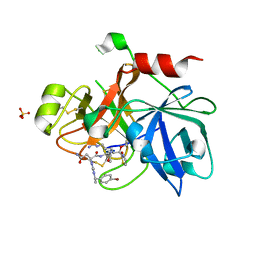 | | Factor XI catalytic domain complexed with N-((R)-1-(4-bromophenyl)ethyl)urea-Asn-Val-Arg-alpha-ketothiazole | | Descriptor: | Coagulation factor XI, N~2~-({[(1R)-1-(4-BROMOPHENYL)ETHYL]AMINO}CARBONYL)ASPARAGINYL-N~1~-{4-{[AMINO(IMINO)METHYL]AMINO}-1-[2,3-DIHYDRO-1,3-THIAZOL-2-YL(HYDROXY)METHYL]BUTYL}VALINAMIDE, SULFATE ION | | Authors: | Lin, J, Deng, H, Jin, L, Prandey, P, Rynkiewicz, M.J, Bibbins, F, Cantin, S, Quinn, J, Magee, S, Gorga, J. | | Deposit date: | 2005-05-18 | | Release date: | 2006-05-09 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Design, Synthesis and Biological Evaluation of Peptidomimetic FXIa Inhibitors
To be Published
|
|
2N5C
 
 | | Solution NMR structure of the lasso peptide chaxapeptin | | Descriptor: | chaxapeptin | | Authors: | Elsayed, S.S, Trusch, F, Deng, H, Raab, A, Prokes, I, Busarakam, K, Asenjo, J.A, Andrews, B.A, van West, P, Bull, A.T, Goodfellow, M, Yi, Y, Ebel, R, Jaspars, M, Rateb, M.E. | | Deposit date: | 2015-07-14 | | Release date: | 2015-10-07 | | Last modified: | 2024-11-20 | | Method: | SOLUTION NMR | | Cite: | Chaxapeptin, a Lasso Peptide from Extremotolerant Streptomyces leeuwenhoekii Strain C58 from the Hyperarid Atacama Desert.
J.Org.Chem., 80, 2015
|
|
3VCB
 
 | | C425S mutant of the C-terminal cytoplasmic domain of non-structural protein 4 from mouse hepatitis virus A59 | | Descriptor: | RNA-directed RNA polymerase | | Authors: | Xu, X, Lou, Z, Ma, Y, Chen, X, Yang, Z, Tong, X, Zhao, Q, Xu, Y, Deng, H, Bartlam, M, Rao, Z. | | Deposit date: | 2012-01-03 | | Release date: | 2012-01-11 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Crystal structure of the C-terminal cytoplasmic domain of non-structural protein 4 from mouse hepatitis virus A59.
Plos One, 4, 2009
|
|
3VC8
 
 | | Crystal structure of the C-terminal cytoplasmic domain of non-structural protein 4 from mouse hepatitis virus A59 | | Descriptor: | RNA-directed RNA polymerase | | Authors: | Xu, X, Lou, Z, Ma, Y, Chen, X, Yang, Z, Tong, X, Zhao, Q, Xu, Y, Deng, H, Bartlam, M, Rao, Z. | | Deposit date: | 2012-01-03 | | Release date: | 2012-01-11 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Crystal structure of the C-terminal cytoplasmic domain of non-structural protein 4 from mouse hepatitis virus A59.
Plos One, 4, 2009
|
|
3J29
 
 | | Dissecting the in vivo assembly of the 30S ribosomal subunit reveals the role of RimM | | Descriptor: | 16S rRNA | | Authors: | Guo, Q, Goto, S, Chen, Y, Muto, A, Himeno, H, Deng, H, Lei, J, Gao, N. | | Deposit date: | 2012-09-28 | | Release date: | 2013-01-16 | | Last modified: | 2024-03-20 | | Method: | ELECTRON MICROSCOPY (14 Å) | | Cite: | Dissecting the in vivo assembly of the 30S ribosomal subunit reveals the role of RimM and general features of the assembly process
Nucleic Acids Res., 41, 2013
|
|
3J2F
 
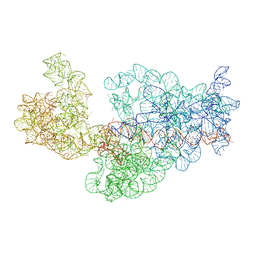 | | Dissecting the in vivo assembly of the 30S ribosomal subunit reveals the role of RimM | | Descriptor: | 16S rRNA | | Authors: | Guo, Q, Goto, S, Chen, Y, Muto, A, Himeno, H, Deng, H, Lei, J, Gao, N. | | Deposit date: | 2012-09-28 | | Release date: | 2013-01-16 | | Last modified: | 2024-03-20 | | Method: | ELECTRON MICROSCOPY (17.6 Å) | | Cite: | Dissecting the in vivo assembly of the 30S ribosomal subunit reveals the role of RimM and general features of the assembly process.
Nucleic Acids Res., 41, 2013
|
|
3J2H
 
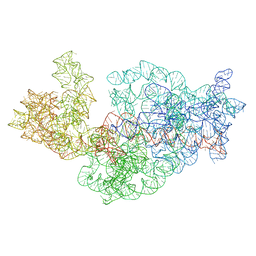 | | Dissecting the in vivo assembly of the 30S ribosomal subunit reveals the role of RimM | | Descriptor: | 16S rRNA | | Authors: | Guo, Q, Goto, S, Chen, Y, Muto, A, Himeno, H, Deng, H, Lei, J, Gao, N. | | Deposit date: | 2012-09-28 | | Release date: | 2013-01-16 | | Last modified: | 2024-03-20 | | Method: | ELECTRON MICROSCOPY (18.799999 Å) | | Cite: | Dissecting the in vivo assembly of the 30S ribosomal subunit reveals the role of RimM and general features of the assembly process.
Nucleic Acids Res., 41, 2013
|
|
3J2E
 
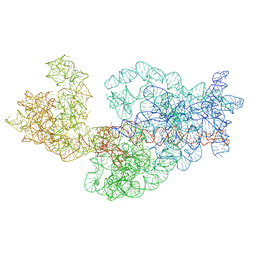 | | Dissecting the in vivo assembly of the 30S ribosomal subunit reveals the role of RimM | | Descriptor: | 16S rRNA | | Authors: | Guo, Q, Goto, S, Chen, Y, Muto, A, Himeno, H, Deng, H, Lei, J, Gao, N. | | Deposit date: | 2012-09-28 | | Release date: | 2013-01-16 | | Last modified: | 2024-03-20 | | Method: | ELECTRON MICROSCOPY (15.3 Å) | | Cite: | Dissecting the in vivo assembly of the 30S ribosomal subunit reveals the role of RimM and general features of the assembly process
Nucleic Acids Res., 41, 2013
|
|
3J2D
 
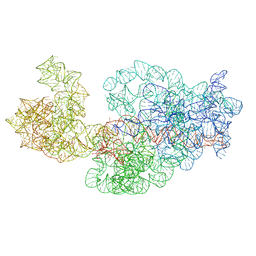 | | Dissecting the in vivo assembly of the 30S ribosomal subunit reveals the role of RimM | | Descriptor: | 16S rRNA | | Authors: | Guo, Q, Goto, S, Chen, Y, Muto, A, Himeno, H, Deng, H, Lei, J, Gao, N. | | Deposit date: | 2012-09-28 | | Release date: | 2013-01-16 | | Last modified: | 2024-03-20 | | Method: | ELECTRON MICROSCOPY (18.700001 Å) | | Cite: | Dissecting the in vivo assembly of the 30S ribosomal subunit reveals the role of RimM and general features of the assembly process
Nucleic Acids Res., 41, 2013
|
|
3J2C
 
 | | Dissecting the in vivo assembly of the 30S ribosomal subunit reveals the role of RimM | | Descriptor: | 16S rRNA body domain, 16S rRNA head domain | | Authors: | Guo, Q, Goto, S, Chen, Y, Muto, A, Himeno, H, Deng, H, Lei, J, Gao, N. | | Deposit date: | 2012-09-28 | | Release date: | 2013-01-16 | | Last modified: | 2024-03-20 | | Method: | ELECTRON MICROSCOPY (13.2 Å) | | Cite: | Dissecting the in vivo assembly of the 30S ribosomal subunit reveals the role of RimM and general features of the assembly process
Nucleic Acids Res., 41, 2013
|
|
