5DIK
 
 | | Crystal structure of apo-lpg0406, a carboxymuconolactone decarboxylase family protein from Legionella pneumophila | | Descriptor: | Alkyl hydroperoxide reductase AhpD | | Authors: | Chen, X, Gong, X, Zhang, N, Ge, H. | | Deposit date: | 2015-09-01 | | Release date: | 2015-10-14 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Structure of lpg0406, a carboxymuconolactone decarboxylase family protein possibly involved in antioxidative response from Legionella pneumophila
Protein Sci., 24, 2015
|
|
8ZQ8
 
 | |
7AUD
 
 | |
7AUC
 
 | |
8ZBF
 
 | |
5XFS
 
 | |
6Z2F
 
 | |
7F8L
 
 | |
8QH2
 
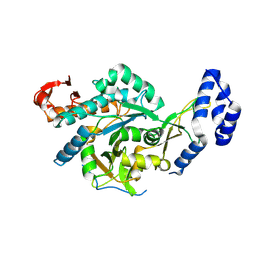 | | Crystal structure of chimeric UAP1L1 | | Descriptor: | UDP-N-acetylhexosamine pyrophosphorylase,UDP-N-acetylhexosamine pyrophosphorylase-like protein 1 | | Authors: | Chen, X, Yan, K, Bartual, S, van Aalten, D. | | Deposit date: | 2023-09-06 | | Release date: | 2024-09-18 | | Last modified: | 2024-11-13 | | Method: | X-RAY DIFFRACTION (2.75 Å) | | Cite: | UAP1L1 is an active pyrophosphorylase with an active site disulfide regulating protein thermostability
To Be Published
|
|
3FUS
 
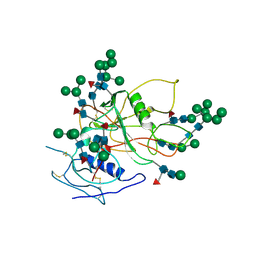 | | Improved Structure of the Unliganded Simian Immunodeficiency Virus gp120 Core | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-[beta-L-fucopyranose-(1-6)]2-acetamido-2-deoxy-beta-D-glucopyranose, ... | | Authors: | Chen, X, Poon, B, Wang, Q, Ma, J. | | Deposit date: | 2009-01-14 | | Release date: | 2009-06-30 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (4 Å) | | Cite: | Structural improvement of unliganded simian immunodeficiency virus gp120 core by normal-mode-based X-ray crystallographic refinement.
Acta Crystallogr.,Sect.D, 65, 2009
|
|
6RWG
 
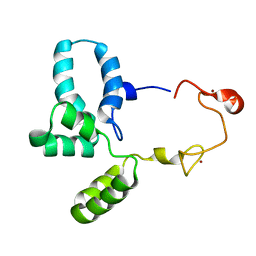 | | Structure of HIV-1 CAcSP1NC mutant(W41A,M42A) interacting with maturation inhibitor EP39 | | Descriptor: | Gag polyprotein, ZINC ION | | Authors: | Chen, X, Coric, P, Larue, V, Nonin-Lecomte, S, Bouaziz, S, Structural Genomics Consortium (SGC) | | Deposit date: | 2019-06-05 | | Release date: | 2020-05-20 | | Last modified: | 2024-06-19 | | Method: | SOLUTION NMR | | Cite: | The HIV-1 maturation inhibitor, EP39, interferes with the dynamic helix-coil equilibrium of the CA-SP1 junction of Gag.
Eur.J.Med.Chem., 204, 2020
|
|
305D
 
 | |
306D
 
 | |
3EJJ
 
 | | Structure of M-CSF bound to the first three domains of FMS | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Colony stimulating factor-1, Macrophage colony-stimulating factor 1 receptor | | Authors: | Chen, X, Liu, H, Focia, P.J, Shim, A, He, X. | | Deposit date: | 2008-09-18 | | Release date: | 2008-12-09 | | Last modified: | 2024-11-20 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Structure of macrophage colony stimulating factor bound to FMS: diverse signaling assemblies of class III receptor tyrosine kinases.
Proc.Natl.Acad.Sci.USA, 105, 2008
|
|
304D
 
 | |
1XCA
 
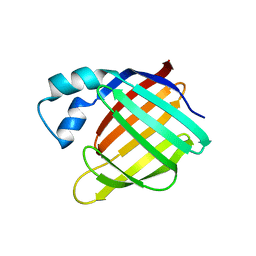 | | APO-CELLULAR RETINOIC ACID BINDING PROTEIN II | | Descriptor: | CELLULAR RETINOIC ACID BINDING PROTEIN TYPE II | | Authors: | Chen, X, Ji, X. | | Deposit date: | 1996-12-31 | | Release date: | 1998-07-01 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Crystal structure of apo-cellular retinoic acid-binding protein type II (R111M) suggests a mechanism of ligand entry.
J.Mol.Biol., 278, 1998
|
|
216D
 
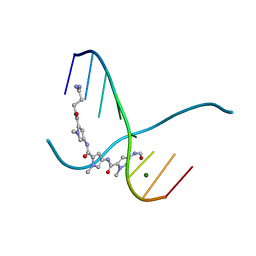 | |
217D
 
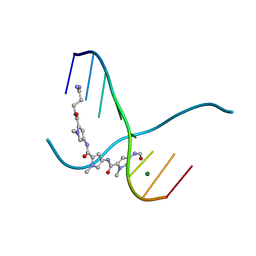 | |
1EGA
 
 | | CRYSTAL STRUCTURE OF A WIDELY CONSERVED GTPASE ERA | | Descriptor: | PROTEIN (GTP-BINDING PROTEIN ERA), SULFATE ION | | Authors: | Chen, X, Ji, X. | | Deposit date: | 1998-12-01 | | Release date: | 1999-07-12 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Crystal structure of ERA: a GTPase-dependent cell cycle regulator containing an RNA binding motif.
Proc.Natl.Acad.Sci.USA, 96, 1999
|
|
1J51
 
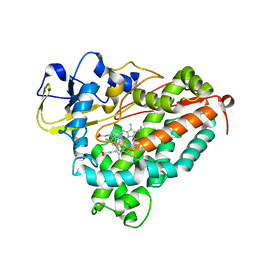 | | CRYSTAL STRUCTURE OF CYTOCHROME P450CAM MUTANT (F87W/Y96F/V247L/C334A) WITH 1,3,5-TRICHLOROBENZENE | | Descriptor: | 1,3,5-TRICHLORO-BENZENE, CYTOCHROME P450CAM, POTASSIUM ION, ... | | Authors: | Chen, X, Christopher, A, Jones, J, Guo, Q, Xu, F, Cao, R, Wong, L.L, Rao, Z. | | Deposit date: | 2002-01-05 | | Release date: | 2002-01-23 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Crystal structure of the F87W/Y96F/V247L mutant of cytochrome P-450cam with 1,3,5-trichlorobenzene bound and further protein engineering for the oxidation of pentachlorobenzene and hexachlorobenzene
J.BIOL.CHEM., 277, 2002
|
|
1JX6
 
 | | CRYSTAL STRUCTURE OF LUXP FROM VIBRIO HARVEYI COMPLEXED WITH AUTOINDUCER-2 | | Descriptor: | 3A-METHYL-5,6-DIHYDRO-FURO[2,3-D][1,3,2]DIOXABOROLE-2,2,6,6A-TETRAOL, CALCIUM ION, LUXP PROTEIN | | Authors: | Chen, X, Schauder, S, Potier, N, Van Dorsselaer, A, Pelczer, I, BassleR, B.L, Hughson, F.M. | | Deposit date: | 2001-09-05 | | Release date: | 2002-02-04 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | Structural identification of a bacterial quorum-sensing signal containing boron.
Nature, 415, 2002
|
|
1KIL
 
 | | Three-dimensional structure of the complexin/SNARE complex | | Descriptor: | Complexin I SNARE-complex binding region, MAGNESIUM ION, SNAP-25 C-terminal SNARE motif, ... | | Authors: | Chen, X, Tomchick, D, Kovrigin, E, Arac, D, Machius, M, Sudhof, T.C, Rizo, J. | | Deposit date: | 2001-12-03 | | Release date: | 2002-03-13 | | Last modified: | 2023-08-16 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Three-dimensional structure of the complexin/SNARE complex.
Neuron, 33, 2002
|
|
6LBF
 
 | | Crystal structure of FEM1B | | Descriptor: | Protein fem-1 homolog B, SULFATE ION | | Authors: | Chen, X, Liao, S, Xu, C. | | Deposit date: | 2019-11-14 | | Release date: | 2020-10-21 | | Last modified: | 2024-03-27 | | Method: | X-RAY DIFFRACTION (3.252 Å) | | Cite: | Molecular basis for arginine C-terminal degron recognition by Cul2 FEM1 E3 ligase.
Nat.Chem.Biol., 17, 2021
|
|
6LDP
 
 | | Structure of CDK5R1-bound FEM1C | | Descriptor: | Protein fem-1 homolog C,Peptide from Cyclin-dependent kinase 5 activator 1, SULFATE ION | | Authors: | Chen, X, Liao, S, Xu, C. | | Deposit date: | 2019-11-22 | | Release date: | 2020-10-21 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (2.35 Å) | | Cite: | Molecular basis for arginine C-terminal degron recognition by Cul2 FEM1 E3 ligase.
Nat.Chem.Biol., 17, 2021
|
|
6LEN
 
 | | Structure of NS11 bound FEM1C | | Descriptor: | Protein fem-1 homolog C,NS11 peptide | | Authors: | Chen, x, Liao, S, Xu, C. | | Deposit date: | 2019-11-25 | | Release date: | 2020-10-21 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (2.383 Å) | | Cite: | Molecular basis for arginine C-terminal degron recognition by Cul2 FEM1 E3 ligase.
Nat.Chem.Biol., 17, 2021
|
|
